Abstract
One of the most important crops worldwide is wheat. Wheat domestication took place about 10,000 years ago. Not only that its wild progenitors have been discovered and phenotypically characterized, but their genomes were also sequenced and compared to modern wheat. While comparative genomics is essential to track genes that contribute to improvement in crop yield, comparative analyses of functional biological end-products, such as metabolites, are still lacking. With the advent of rigorous mass-spectrometry technologies, it is now possible to address that problem on a big-data scale. In attempt to reveal classes of metabolites, which are associated with wheat domestication, we analyzed the metabolomes of wheat kernel samples from various wheat lines. These wheat lines represented subspecies of tetraploid wheat along primary and secondary domestications, including wild emmer, domesticated emmer, landraces durum, and modern durum. We detected that the groups of plant metabolites such as plant-defense metabolites, antioxidants and plant hormones underwent significant changes during wheat domestication. Our data suggest that these metabolites may have contributed to the improvement in the agricultural fitness of wheat. Closer evaluation of specific metabolic pathways may result in the future in genetically-engineered high-yield crops.
Similar content being viewed by others
Introduction
To feed the rapidly growing global population, the scientific community invests intensive efforts in improving the yield and nutritional values of crops, among which is the modern wheat1,2 Modern agriculture technologies and genetic modification techniques are being constantly improved3. Agricultural technology advancement aside, it is crucial to identify which crop traits should be improved, and how to specifically and effectively target them. One of the ways to face that challenge is to investigate crop evolution and domestication. This 10,000-years-long process has eventually resulted in wheat strains far more adapted for human needs, including larger grain size, increase grain number, grain retention and thresh ability required for efficient harvest, high yield, altered photoperiod sensitivity, and a higher glycemic load1,2. These phenotypic traits are a collective endpoint result of a highly complex regulatory genetic network. In between, numerous proteins and metabolites carry out their biological function, creating a vast and diverse biochemical network. Thus, the analytical measurement of these bioactive molecules and their intermediates enables us to functionally link between the plant genotype and the plant phenotype. In the recent decade, analytic “Omics” approaches have become more popular to compare between two or more physiological conditions, between different tissues or between species, consequently producing extensive datasets. Not only that the genomes of modern bread wheat (Triticum aestivum) and durum wheat strains (Triticum turgidum ssp. durum) have been sequenced4,5,6, but also the genomes of their progenitors, including domesticated emmer (Triticum turgidum ssp. dicoccum) and wild emmer (Triticum turgidum ssp. dicoccoides), have been sequenced and compared7,8,9. Many quantitative trait loci and genetic polymorphism markers, which distinguish between modern and wild wheat strains, have been identified and characterized1,4,6,10,11,12. While comparative studies, which utilize genomics, transcriptomics, or proteomics, are quite common13, mass-spectrometry (MS)-coupled metabolomics is not commonly used in crop sciences14. Nevertheless, metabolomics expand our knowledge about how evolutionary genetics during wheat domestication is translated into changes in fluxes of metabolites found in plants14,15. Several recent studies have employed this methodology, for example, to compare between wheat lines16,17, or to evaluate environmental stress-response in wheat18,19,20. Employment of metabolomics to investigate wheat domestication is still uncommon. So far, three metabolomics-based studies have compared between the tetraploid wild emmer, emmer and durum wheat strains: one study analyzed the root exudates21, the second study analyzed the kernels22,23, the third study analyzed changes in specific group of metabolites playing important role in protection of the seeds from a variety of biotic stresses24. In this study we compared ancestor wheat strain wild emmer (WEW, Triticum turgidum ssp. dicoccoides) with its early domesticated version (DEW, Triticum turgidum ssp. dicoccum) and later selected durum wheat (Triticum turgidum ssp. durum), classified into ancient landraces of durum (LD) which are traditional crops, and modern durum (MD) representing high-yielding industrial crops. All those strains are tetraploides. We did not include in this study hexaploid modern bread wheat lines (Triticum aestivum) to avoid unnecessary complexity in comparison with the tetraploid strains. We employed MS-coupled metabolomics with respect to wheat domestication, taking it one step further. In comparison with the previous studies we added more variables, by separating the kernel embryo and endosperm, by creating four categories of wheat strains instead of three, and by increasing the pool of identified metabolites.
Materials and methods
Statement on experimental research on plants
The producers granted permission for plant and seed measurements, which were done in situ. All methods and assays performed in this field study comply with national legislation and guidelines of the Sapir Academic College and Weizmann institute of science, Israel.
Wild emmer seeds wheat (T. turgidum ssp. dicoccoides) and the others type accessions used in this study were provided to us with permission from the collection by Israeli stock centers, as following: Wild Cereals gene bank (WCGB)—Prof Eibi Nevo, Institute of Evolution, University of Haifa and Prof Moshe Feldman, the grain wheat bank of Weizmann institute of science, Israel.
Plant material
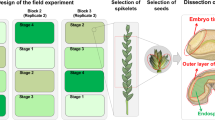
Nineteen wheat lines that correspond to various stages of wheat domestication at the tetraploid level were grown and analyzed (4–5 grains in each line). They consist of wild emmer (WE, Triticum turgidum ssp. dicoccoides), domesticated primitive emmer (PE, Triticum turgidum ssp. dicoccum), domesticated landraces varieties of durum (LD), and modern durum (MD) (Triticum turgidum ssp. durum). The PE and LD lines were collected from traditional farmers, whereas the MD lines represent high-yielding macaroni wheat. The annotations of the wheat lines and their geographical origin are shown in Table 1. The plants were grown under the same conditions, in a net-house with three replicates per line; each replicate being grown in a separate block. All plants were grown in 3-L pots during the winter25.
Sample preparation
Whole grain samples, separated into endosperm and embryo, were used for the study. The metabolite extraction was performed as described previously24. In brief, the embryo and endosperm tissues samples (500 mg) of different lines were ground to a fine powder in liquid nitrogen, and 1.5 mL of 75% methanol with 0.1% formic acid was added. After sonication at room temperature for 15 min, the samples were centrifuged (10,000g) and filtered. The samples were stored at − 20 °C prior to analysis. The metabolites in the extractable fraction were further purified by column chromatography (Amberlite XAD 8 HP) and eluted with ethanol. Both fractions were freeze-dried and stored at − 80 °C.
LC–MS metabolite analysis
Metabolite analysis was carried out using as described by Hanhineva et al. (2008, 2011) and by Ben Abu et al. (2021)24,25. This procedure used a UPLC-PDA-qTOF-MS system: a UPLC Waters Acquity instrument connected in-line to an Acquity PDA (photodiode array) detector and a Synapt HDMS detector (tandem quadrupole/time-of-flight mass spectrometer). This analysis was carried out using ultra-performance liquid chromatography coupled with a photodiode detector-quadrupole and tandem time-of-flight mass spectrometry (UPLC-PDA-qTOF-MS-Waters Premier qTOF, Milford, MA, USA). The system consisted of a Acquity UPLC (Waters) connected in-line to an Acquity PDA detector (Waters) and a Synapt HDMS detector (Waters). The HDMS system was operated in the standard qTOF mode, without using the ion mobility capabilities. Metabolite separation was performed using a UPLC BEH C18 column (100 × 2.1 mm i.d., 1.7 μm; Waters). The mobile phase consisted of 0.1% formic acid in acetonitrile/water (5:95, v/v) (phase A) and 0.1% formic acid in acetonitrile (phase B). The linear gradient program was as follows: 100–72% A over 22 min, 72–60% A over 0.5 min, 60–0% A over 0.5 min, holding at 100% B for a 1.5 min, then returning to initial conditions (100% A) over 0.5 min, and conditioning at 100% A. The flow rate was 0.3 mL/min, and the column temperature was kept at 35 °C. The UV spectra were recorded at 210–550 nm using the Acquity PDA detector (Waters), or the UV trace was measured at 240 nm using the Acquity UV detector (Waters). Eluting compounds were detected using the qTOF equipped with an electrospray ionization (ESI) source. Acquisition was performed in ESI-positive and ESI-negative modes. The following settings were applied during the LC–MS runs: capillary voltage, 3.0 kV; cone voltage, 30 eV; collision energy, 3 eV and 20 eV; and collision gas, argon. For the LCMS/MS analysis, collision energies of 20 and 35 eV were used. The m/z range was 50–1500 Da. The MS was calibrated using sodium formate, and leucine enkephalin was used as the lock mass. A standard mixture containing 40 μg/mL of each of the following compounds was used to monitor the quality of the chromatogram, to ensure the consistency of retention times across runs, and to aid in metabolite identification: L-tryptophan, L-phenylalanine, p-coumaric acid, caffeic acid, sinapic acid, benzoic acid, quercetin dehydrate, kaempferol, rutin, and trans-resveratrol (all purchased from Sigma); naringenin, chlorogenic acid hemihydrate, trans-cinnamic acid, and isorhamnetin (Fluka); ferulic acid (Aldrich); and tomatine (Apin chemicals). Mass Lynx v4.1 (Waters) was used to control all instruments and to calculate accurate masses7,26,27,28,29,30.
LC–MS data analysis
The chromatograms obtained from UPLC PDA-qTOF-MS analysis were processed by the Marker Lynx 4.1 software (Waters) for mass signal extraction and alignment. The identification of metabolites was performed as described previously on data obtained using the ESI-negative mode24. In short, the accurate mass and molecular formula predictions were screened for putative molecules from the Dictionary of Natural Products (Chapman and Hall/ CRC) and the SciFinder Scholar databases (SciFinder Scholar 2007) as described elsewhere25. The MS/MS fragmentation and UV-absorbability of the metabolites were compared with those of candidate molecules found in databases and verified with earlier literature on similar compounds23.
Software and statistical analysis
Dataset and statistical analyses were done using Microsoft Excel 365, MATLAB 8.0 and Statistics Toolbox 8.1 software. p (FDR) < 0.05 is considered significant. We ascribed function to metabolites based on databases, and complementary internet search. Figures were generated using, MATLAB 8.0, GraphPad Prism 7.0 and Meta-Chart.
Results
Identification of metabolites associated with wheat domestication
We collected wild, primitive, and domesticated tetraploid wheat lines form various eco-geographical locations (Table 1). In order to associate metabolite profiling with wheat domestication, we classified these wheat lines into four categories (from wild lines to most domesticated lines): WEW, DEW, LD, MD. These wheat lines belong to three taxonomic subspecies of tetraploid Triticum turgidum, representing primary (WEW to DEW) and secondary (DEW to LD/MD) domestications1. To analyze specific trends, we identified metabolites present in wheat kernels using an annotated database24. In the embryo samples, we identified 580 metabolites, whereas in the endosperm samples, we identified 89 metabolites. We next attempted to characterize which metabolites undergo changes in association with wheat domestication. For that purpose, we undertook used PCA analysis as we perform elsewhere25 and appear in Fig. 1A,B. Moreover, we used two statistical approaches. First, we directly evaluated which identified metabolites significantly increase or decrease when the LD and MD groups are compared with the WEW group. In the embryo kernel, a total of 47.9% showed an increase, and a total of 13.5% showed a decrease (Fig. 2A—small pie chart). In the kernel endosperm, a total of 31.5% showed an increase, and a total of 21.4% showed a decrease (Fig. 2B—small pie chart). Secondly, we evaluated which metabolites steadily increase or decrease in both primary and secondary domestications (i.e., WEW to DEW and DEW to LD/MD respectively). Our analysis revealed that out of the identified metabolites in the kernel embryo 7.4% increased and 3.8% decreased in association with wheat domestication (Fig. 2A—large pie chart, Table S1). In the kernel endosperm, 1.1% increased, and 2.3% decreased in association with wheat domestication (Fig. 2B—large pie chart, Table S1). A very few metabolites remained conserved in the kernel embryo, while a relatively higher proportion of metabolites remained conserved in the kernel endosperm (Fig. 2A,B—large pie charts, Table S1). Breaking apart primary and secondary domestications revealed that many more metabolites underwent changes in the domestication steps considered here (Fig. 2C,D), therefore only a small portion of metabolites steadily changed over time (Fig. 2A,D). All the other metabolites were variable in their expression behavior. Together, these results suggest that the kernel embryo was subjected to a greater evolutionary pressure upon domestication. Considering the larger pool of metabolites detected in the embryo and the observation that metabolites in the endosperm did not undergo marked changes (fold-change-wise, not shown), we further focused on the embryo.
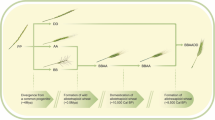
Three-dimensional models of principal component analyses (PCA). Each ball represents a quantile-normalized metabolome of one wheat line replica sample. (A) Embryo metabolomes—two perspectives. (B) Endosperm metabolomes—two perspectives. The dashed circles surround distinct groups of wheat metabolomes. WEW—wild emmer; WED (PE)—primitive emmer; LD—landraces durum; MD—modern durum. (C). Evolutionary history of allotetraploid and allohexaploid wheat: Diploid wheats (2n = 2 × = 14), from the Tritcum-Aegilops group have diverged ~ 4Mya from a diploid progenitor whose genome is indicated here as PP. Intergeneric hybridization between the diploid T. urartu (genome AA) as male and the donor of BB genome as female, (an unknown species similar to Ae. speltoides), followed by chromosome doubling, gave rise (~ 0.5Mya) to the wild allotetraploid wheat, Triticum turgidum, ssp. dicoccoides (genome BBAA, 2n = 4 × = 28), the direct progenitor of durum and bread wheat. Domestication of allotetraploid wheat took place ~ 10,500 years ago and was followed by a second round of intergeneric hybridization chromosome doubling between domesticated allotetraploid wheat and the donor of the D genome, Ae. Tauschii (2n = 2 × = 14, genome DD), giving rise, ~ 9000 years ago, to bread wheat, an allohexaploid (2n = 6 × = 42, genome AABBDD).
Comparative metabolomics. Identified metabolites that underwent significant changes in association with wheat domestication. Each metabolome is a result of group-averaged metabolite values from all the replicas and wheat lines that belong to that group. A statistical comparison is made between each pair of groups from a total of 4 groups: WEW—wild emmer; WED (PE)—primitive emmer; LD—landraces durum; MD—modern durum. (A,C) Comparison of embryo metabolomes. (B,D) Comparison of endosperm metabolomes. (A-B) Small pie charts—comparisons between the WE group and the LD/MD groups. Metabolites that underwent a significant increase or decrease in two comparisons (WE to LD and WE to MD). Variable metabolites either did not undergo statistically significant changes or did not increase or decrease in both comparisons. Large pie charts—comparisons between the WE, PE group and the LD/MD groups. Metabolites that underwent a significant increase or decrease in three comparisons (WE to PE, PE to LD and PE to MD) were considered to be steadily changed metabolites. Conserved metabolites did not undergo any statistical changes in any comparison. Variable metabolites did not undergo a steady increase or a steady decrease in all three comparisons. (C-D) Venn diagrams depict the numbers of statistically changed metabolites in the following comparisons: WE to PE, PE to both LD and MD, and WE to both LD and MD, and steadily changed metabolites. Increased metabolites on the left and decreased metabolites on the right.
Heat-maps show how certain metabolites in the kernel embryo, classified into 4 clusters, were co-regulated among wheat lines within the same group (Fig. S1), suggesting common co-regulated metabolic pathways. It is reasonable to assume that upon wheat domestication, the evolutionary pressure selected for certain biological functions and suppressed others. Thus, we classified the significantly changed metabolites into categories of biological function, using online databases (see “Materials and methods” section). Strikingly, the top-ranked category of increased metabolites consisted of compounds that are known to be involved directly or indirectly in plant defense mechanisms. This category comprised 23.4% of the increased metabolites (see also Fig. 2A—large pie chart) and 41.9% of the steadily increased metabolites (see also Fig. 2A—small pie chart). These compounds included phytoalexins, jasmonic acids, benzoxazinoids, glucosinolates, allelochemicals, coumaric acids, and others (Fig. 3A). The second and third top-ranked categories consisted of antioxidants and metabolites that are involved in growth and development (20.5% and 12.9% of increased metabolites, respectively). These compounds included cinnamic acid derivatives, apigenin derivatives, flavonols, glutathiones, anthocyanins, and several plant hormones (Fig. 3B,C). As for significantly decreased metabolites, we did not identify any outstanding category based on biological function. Metabolites that participate in amino acid metabolism was also evident among increased metabolites. The relative composition of amino acids in wheat kernels was therefore altered with domestication; we detected an increase in half of the amino acids and a decrease in one amino acid (Fig. 3D). In order to identify additional metabolites of interest, we generated six volcano plots, which took into account the fold-change and the statistical significance. Each volcano plot compares between a pair of group-averaged embryo metabolomes (Fig. 4A-F). The metabolites that underwent the most significant alterations are shown in Table S2 (also marked in Fig. 4A-F). Altogether, our findings demonstrated that upon wheat domestication, the wheat metabolome had been substantially altered. Follow-up research will determine how altered metabolic pathways and specific metabolites contributed to the fitness of modern wheat to agricultural needs.
Categories of biological function. Embryo metabolomes. Identified metabolites that underwent a significant increase in association with wheat domestication (WE to LD and MD) were classified according to chemical structure and biological function. (A) Metabolites that are involved in plant defense mechanisms. (B) Metabolites that are known to be antioxidants. (C) Plant hormones and their intermediates. (D) Proteinogenic amino acids.
Volcano plots were generated based on comparative metabolomics between the average embryo metabolomes of each group (only identified metabolites, see also Fig. 3). WEW—wild emmer; DEW—primitive emmer; LD—landraces durum; MD—modern durum. All the metabolites that are above the horizontal line of p < 0.05 were considered significant. Red rectangles insets—the most significantly changed metabolites are shown in Tables S1 and S2.
Discussion
In this study, we employed MS-coupled metabolomics to compare between the metabolomes of tetraploid wheat lines, classified in 4 groups that follow the course of wheat domestication (Fig. 1C). The primary domestication is the evolution of WEW to DEW, whereas the secondary domestication is the evolution of DEW to durum wheat1, which is also divided into two groups—LD (i.e., traditional) and MD (i.e., industrial). Three-dimensional (3D) principal component analyses (PCAs), run without a wheat category-bias, also demonstrated a clear separation among the WEW, DEW, and durum accessions in both the embryo and the endosperm tissues as describe here and elsewhere y Ben Abu and Itsko25 (Fig. 1). Thus, the heatmaps and the PCAs were consistent with our hypothesis that metabolite composition and expression underwent substantial changes during wheat domestication. Our comparative analysis revealed that many metabolites in wheat kernels had undergone significant changes in each step of domestication. Here, we focused on the specific metabolites of the kernel embryo which show the same tendency of changes during both primary and secondary event of domestication. While many metabolites underwent significant changes, fewer metabolites steadily increased or decreased in association with both domestications events. The output of our comparative metabolomics revealed a multitude of metabolites that were not discussed previously in former metabolomic studies of wheat domestication21,22. Notably, we detected a substantial increase in the expression of secondary metabolites that participate in plant-defense mechanisms and response to biotic stress. These compounds include phytoalexins, jasmonic acids, benzoxazinoids, glucosinolates, coumaric acids, and other alkaloids that serve as bactericides, fungicides, and insecticides30,31,32,33. In a previous report, we characterized the stress response of WEW by producing benzoxazinoids15,25. We also detected herbicidal allelochemicals that inhibit the growth of surrounding plants25,34. Disease-resistance is one of the most dominant adaptive traits in crop productivity35, however, the acquirement of disease-resistance qualities during wheat domestication has not been fully investigated36. Few reports have compared thereof the resistance to specific pathogens37,38. Interestingly, some of these defense-related metabolites were downregulated when MD lines are compared with LD lines. It is possible that the use of industrial pesticides has led to a loss in the expression of these endogenous pesticides, but not to the extent they are normalized down to the levels found in wild plants. Thus, it could be valuable to improve crop resistance, while minimizing dependence on harmful industrial pesticides39. Genetic engineering of crops that promotes the release of phytochemicals that confer resistance to biotic stress may be the technological solution. Genetic regulation of disease resistance in plants is a field of extensive research, however, scientists still need to functionally link between the genes and the bioactive metabolites, so as to genetically improve wheat strains. Several studies have identified certain genes in wheat that promote the expression of defense-mediating metabolites, including metabolites that we associated with wheat domestication40,41,42. Both biotic stress (e.g., pathogens) and abiotic stress (e.g., lack of nutrients, extreme temperatures, and drought) may lead to oxidative stress in plants, against which antioxidants relay protection43. Our comparative metabolomics revealed that many antioxidants accumulate in the wheat kernels as an adaptive trait that the domesticated crop most likely acquired. These compounds include cinnamic acid derivatives, apigenin derivatives, flavonols, glutathiones, anthocyanins, and other phenols43. Genes that induce expression of antioxidants in wheat may also confer disease resistance44,45. Apart from benefits for wheat fitness, high nutritional values and health benefits are attributes of high antioxidant concentrations in plants as a food source46,47,48. Agricultural yield depends on fast and sustained growth of crops under a broad range of environmental conditions as well as on the development of many-fold large grains49. The basis for the “green revolution” in the last decades was the introduction of genes that increase crop productivity via phytohormonal regulation50. More recent genomic research aims at characterizing the genetic network that contributes to wheat productivity51,52,53,54. Thus, it is of no surprise that we detected an elevation in plant hormones and their intermediates in domesticated wheat as compared with its primitive and wild progenitors. Additionally, we noticed that amino acid metabolism was significantly altered in the course of wheat domestication, demonstrating an increased proportion of several amino acids. Interestingly only one amino acid (tyrosine) demonstrated decrease during domestication (Fig. 3). Tyrosine is the direct precusor of p-coumaric acid which is a component of defence mechanisms of wheat against biotic and abiotic stresses. Therefore, the relative decrease in level of tyrosine may indicate increased metabolic flux from its pool to that one of p-coumaric acid.
In summary, our metabolomic data can be helpful to identify novel genes that promote the synthesis of defense-related metabolites, antioxidants, and plant hormones; not only in the context of with wheat domestication, but also in the context of genetic engineering of high-yield wheat strains.
Data availiability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Haas, M., Schreiber, M. & Mascher, M. Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. J. Integr. Plant Biol. 61(3), 204–225 (2019).
Hebelstrup, K. H. Differences in nutritional quality between wild and domesticated forms of barley and emmer wheat. Plant Sci. 256, 1–4 (2017).
Borisjuk, N. et al. Genetic modification for wheat improvement: From transgenesis to genome editing. Biomed. Res. Int. 2019, 6216304 (2019).
Maccaferri, M. et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51(5), 885–895 (2019).
Brenchley, R. et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491(7426), 705–710 (2012).
Zimin, A. V. et al. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6(11), 1–7 (2017).
Avni, R. et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357(6346), 93–97 (2017).
Luo, M. C. et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551(7681), 498–502 (2017).
Jia, J. et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496(7443), 91–95 (2013).
Peng, J. et al. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. U. S. A. 100(5), 2489–2494 (2003).
Allen, A. M. et al. Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol. J. 11(3), 279–295 (2013).
Merchuk-Ovnat, L., Fahima, T., Krugman, T. & Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve grain yield, biomass and photosynthesis across enviroinments in modern wheat. Plant Sci. 251, 23–34 (2018).
Bhalla, P. L., Sharma, A. & Singh, M. B. Enabling molecular technologies for trait improvement in wheat. Methods Mol. Biol. 1679, 3–24 (2017).
Hong, J., Yang, L., Zhang, D., & Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 17(6), 1–16 (2016).
Batyrshina, Z. S., Yaakov, B., Shavit, R., Singh, A. & Tzin, V. Comparative transcriptomic and metabolic analysis of wild and domesticated wheat genotypes reveals differences in chemical and physical defense responses against aphids. BMC Plant Biol. 20(1), 19 (2020).
Zorb, C., Langenkamper, G., Betsche, T., Niehaus, K. & Barsch, A. Metabolite profiling of wheat grains (Triticum aestivum L.) from organic and conventional agriculture. J. Agric. Food Chem. 54(21), 8301–8306 (2006).
Matthews, S. B. et al. Metabolite profiling of a diverse collection of wheat lines using ultraperformance liquid chromatography coupled with time-of-flight mass spectrometry. PLoS ONE 7(8), e44179 (2012).
de Leonardis, A. M. et al. Effects of heat stress on metabolite accumulation and composition, and nutritional properties of durum wheat grain. Int. J. Mol. Sci. 16(12), 30382–30404 (2015).
Allwood, J. W. et al. Profiling of spatial metabolite distributions in wheat leaves under normal and nitrate limiting conditions. Phytochemistry 115, 99–111 (2015).
Ullah, N., Yuce, M., Neslihan Ozturk Gokce, Z. & Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genom. 18(1), 969 (2017).
Lannucci, A., Fragasso, M., Beleggia, R., Nigro, F. & Papa, R. Evolution of the crop rhizosphere: Impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front Plant Sci. 8, 2124 (2017).
Beleggia, R. et al. Evolutionary metabolomics reveals domestication-associated changes in tetraploid wheat kernels. Mol. Biol. Evol. 33(7), 1740–1753 (2016).
Poudel, R., Bhinderwala, F., Morton, M., Powers, R. & Rose, D. J. Metabolic profiling of historical and modern wheat cultivars using proton nuclear magnetic resonance spectroscopy. Sci. Rep. 11(1), 3080 (2021).
Hanhineva, K. et al. Non-targeted analysis of spatial metabolite composition in strawberry (Fragariaxananassa) flowers. Phytochemistry 69(13), 2463–2481 (2008).
Ben-Abu, Y. & Itsko, M. "Changes in “natural antibiotic” metabolite composition during tetraploid wheat domestication. Sci. Rep. 11(1), 20340. https://doi.org/10.1038/s41598-021-98764-5 (2021).
Salamini, F., Ozkan, H., Brandolini, A., Schäfer-Pregl, R. & Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3(6), 429–441. https://doi.org/10.1038/nrg817 (2002).
Zörb, C., Langenkämper, G., Betsche, T., Niehaus, K. & Barsch, A. Metabolite profiling of wheat grains (Triticum aestivum L.) from organic and conventional agriculture. J. Agric. Food Chem. 54(21), 8301–8306 (2006).
Ben-Abu, Y., Beiles, A., Flom, D. & Nevo, E. Adaptive evolution of benzoxazinoids in wild emmer wheat, Triticum dicoccoides, at “Evolution Canyon”, Mount Carmel, Israel. PLoS ONE. 13(2), e0190424 (2018).
Ben-Abu, Y., et al., Durum wheat evolution—a genomic analysis. In Proceedings of the International Symposium on Genetics and Breeding of Durum Wheat, Vol. 110 29–44 (2014).
Zaynab, M. et al. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 124, 198–202 (2018).
de Bruijn, W. J. C., Gruppen, H. & Vincken, J. P. Structure and biosynthesis of benzoxazinoids: Plant defence metabolites with potential as antimicrobial scaffolds. Phytochemistry 155, 233–243 (2018).
Arbona, V. & Gomez-Cadenas, A. Metabolomics of Disease resistance in crops. Mol. Biol. 19, 13–30 (2016).
Okada, K., Abe, H. & Arimura, G. Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol. 56(1), 16–27 (2015).
Belz, R. G. Allelopathy in crop/weed interactions–an update. Pest. Manag. Sci. 63(4), 308–326 (2007).
Mondal, S. et al. Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front. Plant Sci. 7, 991 (2016).
Huang, L. et al. Evolution and adaptation of wild emmer wheat populations to biotic and abiotic stresses. Annu. Rev. Phytopathol. 54, 279–301 (2016).
Ben-David, R., Dinoor, A., Peleg, Z. & Fahima, T. Reciprocal hosts’ responses to powdery mildew isolates originating from domesticated wheats and their wild progenitor. Front. Plant Sci. 9, 75 (2018).
Yahiaoui, N., Brunner, S. & Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 47(1), 85–98 (2006).
Parween, T., Jan, S., Mahmooduzzafar, S., Fatma, T. & Siddiqui, Z. H. Selective effect of pesticides on plant—a review. Crit. Rev. Food Sci. Nutr. 56(1), 160–179 (2016).
Mou, Y., et al. Genome-wide identification and characterization of the OPR gene family in wheat (Triticum aestivum L). Int. J. Mol. Sci. 20(8), 85–97 (2019).
Kage, U., Karre, S., Kushalappa, A. C. & McCartney, C. Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol. J. 15(4), 447–457 (2017).
Dutartre, L., Hilliou, F. & Feyereisen, R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: Gene duplications and origin of the Bx cluster. BMC Evol. Biol. 12, 64 (2012).
Gill, S. S. & Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48(12), 909–930 (2010).
Dhokane, D., Karre, S., Kushalappa, A. C. & McCartney, C. Integrated metabolo-transcriptomics reveals fusarium head blight candidate resistance genes in wheat QTL-Fhb2. PLoS ONE 11(5), e0155851 (2016).
Kage, U., Yogendra, K. N. & Kushalappa, A. C. TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 7, 42596 (2017).
Masisi, K., Beta, T. & Moghadasian, M. H. Antioxidant properties of diverse cereal grains: A review on in vitro and in vivo studies. Food Chem. 196, 90–97 (2016).
Sova, M. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini Rev. Med. Chem. 12(8), 749–767 (2012).
Perez-Vizcaino, F. & Fraga, C. G. Research trends in flavonoids and health. Arch Biochem. Biophys. 646, 107–112 (2018).
Kong, L., Guo, H. & Sun, M. Signal transduction during wheat grain development. Planta 241(4), 789–801 (2015).
Nadolska-Orczyk, A., Rajchel, I. K., Orczyk, W. & Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor Appl Genet 130(6), 1081–1098 (2017).
Li, W. & Yang, B. Translational genomics of grain size regulation in wheat. Theor. Appl. Genet. 130(9), 1765–1771 (2017).
Qi, P. F. et al. Transcriptional reference map of hormone responses in wheat spikes. BMC Genom. 20(1), 390 (2019).
Hill, C. B. & Li, C. Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Front .Plant Sci. 7, 1906 (2016).
Jiang, Y., Schmidt, R. H., Zhao, Y. & Reif, J. C. A quantitative genetic framework highlights the role of epistatic effects for grain-yield heterosis in bread wheat. Nat. Genet. 49(12), 1741–1746 (2017).
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ben-Abu, Y., Itsko, M. Metabolome dynamics during wheat domestication. Sci Rep 12, 8532 (2022). https://doi.org/10.1038/s41598-022-11952-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11952-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.