Abstract
Nucleoprotein complexes play an integral role in genome organization of both eukaryotes and prokaryotes. Apart from their role in locally structuring and compacting DNA, several complexes are known to influence global organization by mediating long-range anchored chromosomal loop formation leading to spatial segregation of large sections of DNA. Such megabase-range interactions are ubiquitous in eukaryotes, but have not been demonstrated in prokaryotes. Here, using a genome-wide sedimentation-based approach, we found that a transcription factor, Rok, forms large nucleoprotein complexes in the bacterium Bacillus subtilis. Using chromosome conformation capture and live-imaging of DNA loci, we show that these complexes robustly interact with each other over large distances. Importantly, these Rok-dependent long-range interactions lead to anchored chromosomal loop formation, thereby spatially isolating large sections of DNA, as previously observed for insulator proteins in eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Data availability
All raw and processed sequencing datasets generated during this study can be accessed at Gene Expression Omnibus repository under accession number GSE144475.
Code availability
All source code used in this study has been published before and is referenced in the Methods section.
References
Le, T. B. K., Imakaev, M. V., Mirny, L. A. & Laub, M. T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013).
Toro, E. & Shapiro, L. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2, a000349 (2010).
Wang, X., Llopis, P. M. & Rudner, D. Z. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14, 191–203 (2013).
Lioy, V. S. et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783 (2018).
Szabo, Q., Bantignies, F. & Cavalli, G. Principles of genome folding into topologically associating domains. Sci. Adv. 5, eaaw1668 (2019).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Le Gall, A., Valeri, A. & Nollmann, M. Roles of chromatin insulators in the formation of long-range contacts. Nucleus 6, 118–122 (2015).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Parelho, V. et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 (2008).
Brasset, E. & Vaury, C. Insulators are fundamental components of the eukaryotic genomes. Heredity 94, 571–576 (2005).
Rubin, A. J. et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017).
Dixon, J. R., Gorkin, D. U. & Ren, B. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680 (2016).
Le, T. B. & Laub, M. T. Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 35, 1582–1595 (2016).
Dame, R. T., Rashid, F. Z. M. & Grainger, D. C. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 21, 227–242 (2019).
Hirano, T. Condensin-based chromosome organization from bacteria to vertebrates. Cell 164, 847–857 (2016).
Wang, X., Brandão, H. B., Le, T. B. K., Laub, M. T. & Rudner, D. Z. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017).
Lioy, V. S., Junier, I., Lagage, V., Vallet, I. & Boccard, F. Distinct activities of bacterial condensins for chromosome management in Pseudomonas aeruginosa. Cell Rep. 33, 108344 (2020).
Takemata, N., Samson, R. Y. & Bell, S. D. Physical and functional compartmentalization of archaeal chromosomes. Cell 179, 165–179 (2019).
Marbouty, M. et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602 (2015).
Schoenfelder, S. & Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
West, A. G., Gaszner, M. & Felsenfeld, G. Insulators: many functions, many mechanisms. Genes Dev. 16, 271–288 (2002).
Brunwasser-Meirom, M. et al. Using synthetic bacterial enhancers to reveal a looping-based mechanism for quenching-like repression. Nat. Commun. 7, 10407 (2016).
Smirnov, A. et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl Acad. Sci. USA 113, 11591–11596 (2016).
Fernandez-Martinez, J., LaCava, J. & Rout, M. P. Density gradient ultracentrifugation to isolate endogenous protein complexes after affinity capture. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot087957 (2016).
Seid, C. A., Smith, J. L. & Grossman, A. D. Genetic and biochemical interactions between the bacterial replication initiator DnaA and the nucleoid-associated protein Rok in Bacillus subtilis. Mol. Microbiol. 103, 798–817 (2017).
Hoa, T. T., Tortosa, P., Albano, M. & Dubnau, D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43, 15–26 (2002).
Albano, M. et al. The rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 187, 2010–2019 (2005).
Smits, W. K. & Grossman, A. D. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 6, e1001207 (2010).
Duan, B. et al. How bacterial xenogeneic silencer rok distinguishes foreign from self DNA in its resident genome. Nucleic Acids Res. 46, 10514–10529 (2018).
Slager, J., Kjos, M., Attaiech, L. & Veening, J. W. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157, 395–406 (2014).
Karaboja, X. et al. XerD unloads bacterial SMC complexes at the replication terminus. Mol. Cell 81, 756–766 (2021).
Murray, H. & Koh, A. Multiple regulatory systems coordinate DNA replication with cell growth in Bacillus subtilis. PLoS Genet. 10, e1004731 (2014).
Trussart, M. et al. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat. Commun. 8, 14665 (2017).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Kovács, Á. T. & Kuipers, O. P. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J. Bacteriol. 193, 998–1002 (2011).
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009).
Ghirlando, R. & Felsenfeld, G. CTCF: making the right connections. Genes Dev. 30, 881–891 (2016).
Hu, G. et al. Systematic screening of CTCF binding partners identifies that BHLHE40 regulates CTCF genome-wide distribution and long-range chromatin interactions. Nucleic Acids Res. 48, 9606–9620 (2020).
Liu, Z., Scannell, D. R., Eisen, M. B. & Tjian, R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146, 720–731 (2011).
Qiu, Z. et al. Functional interactions between NURF and Ctcf regulate gene expression. Mol. Cell. Biol. 35, 224–237 (2015).
Donohoe, M. E., Zhang, L. F., Xu, N., Shi, Y. & Lee, J. T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell 25, 43–56 (2007).
Guastafierro, T. et al. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J. Biol. Chem. 283, 21873–21880 (2008).
Stik, G. et al. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat. Genet. 52, 655–661 (2020).
Zhang, D. et al. Alteration of genome folding via contact domain boundary insertion. Nat. Genet. 52, 1076–1087 (2020).
Goosen, N. & van de Putte, P. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 16, 1–7 (1995).
Wang, X. et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–75 (2015).
Soh, Y. M. et al. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366, 1129–1133 (2019).
Graham, T. G. W. et al. ParB spreading requires DNA bridging. Genes Dev. 28, 1228–38 (2014).
Funnell, B. E. ParB partition proteins: complex formation and spreading at bacterial and plasmid centromeres. Front. Mol. Biosci. 3, 44 (2016).
Hao, N., Shearwin, K. E. & Dodd, I. B. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat. Commun. 8, 1628 (2017).
Hao, N., Shearwin, K. E. & Dodd, I. B. Positive and negative control of enhancer-promoter interactions by other DNA loops generates specificity and tunability. Cell Rep. 26, 2419–2433 (2019).
Qin, L., Erkelens, A. M., Markus, D. & Dame, R. T. The B. subtilis Rok protein compacts and organizes DNA by bridging. Preprint at bioRxiv https://doi.org/10.1101/769117 (2019).
van der Valk, R. A. et al. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife 6, e27369 (2017).
Chen, J. M. et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 36, 2123–2135 (2008).
Cournac, A. & Plumbridge, J. DNA looping in prokaryotes: experimental and theoretical approaches. J. Bacteriol. 195, 1109–1119 (2013).
Semsey, S., Tolstorukov, M. Y., Virnik, K., Zhurkin, V. B. & Adhya, S. DNA trajectory in the Gal repressosome. Genes Dev. 18, 1898–1907 (2004).
Spizizen, J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl Acad. Sci. USA 44, 1072–1078 (1958).
Jahn, N., Brantl, S. & Strahl, H. Against the mainstream: the membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 98, 651–66 (2015).
Dugar, G. et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 9, e1003495 (2013).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–165 (2016).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Crémazy, F. G. et al. Determination of the 3D genome organization of bacteria using Hi-C. Methods Mol. Biol. 1837, 3–18 (2018).
Wolff, J. et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 46, W11–W16 (2018).
Hofmann, A., Müggenburg, J., Crémazy, F. & Heermann, D. W. Bekvaem: integrative data explorer for Hi-C data. J. Bioinform. Genom. https://doi.org/10.18454/jbg.2019.2.11.1 (2019).
Sinkhorn, R. & Knopp, P. Concerning nonnegative matrices and doubly stochastic matrices. Pacific J. Math. 21, 343–348 (1967).
Wang, X., Llopis, P. M. & Rudner, D. Z. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc. Natl Acad. Sci. USA 111, 12877–12882 (2014).
Vischer, N. O. E. et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front. Microbiol. 6, 586 (2015).
Acknowledgements
We thank R. Dame and F. Z. Rashid for providing critical input on Hi-C protocol, S. van Leeuwen (MAD, University of Amsterdam) for providing excellent sequencing services and N. Vischer for help with FROS image analysis. This work was funded by EMBO, ALTF 936–2016 (G.D.), European Commission MCSA-IF grant no. 749510 (G.D.), Netherlands Organization for Scientific Research (NWO) VENI grant-VI.VENI.192.103 (G.D.) and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy EXC-2181/1–390900948, the Heidelberg STRUCTURES Cluster of Excellence (D.W.H.).
Author information
Authors and Affiliations
Contributions
G.D. conceived the project, designed and performed all experiments (including Hi-C library preparation, analysis and visualization), analyzed data and wrote the manuscript. A.H. and D.W.H. performed Hi-C data analysis and visualization, and provided input on manuscript preparation. L.W.H. conceived the project, analyzed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Marcelo Nollmann, Jesse Dixon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
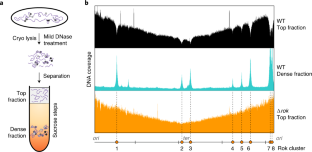
Extended Data Fig. 1 Sedimentation based DNA coverage maps and identification of Rok associated large DNA complexes.
a) Illustration of the expected sedimentation based DNA coverage maps. b) DNA coverage maps from the top fraction of wild type (black) and Δrok (orange) strains. The ratio of top fraction coverage plots (Δrok/wt) shows the Rok clusters as peaks (grey). DNA coverage of DNA obtained from the dense fraction is shown in cyan. Rok clusters are defined at sites where both local minima and local maxima are observed in DNA coverage of top fraction and dense fraction, respectively (Supplementary Table 3). Rok ChIP data in red is also shown along the coverage files.
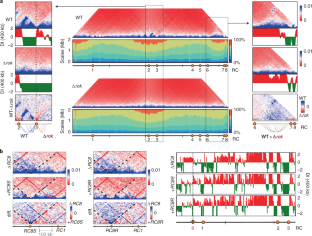
Extended Data Fig. 2 Rok multimerization is essential for interaction between Rok clusters.
a) Normalized Hi-C contact maps of wild type, rok deletion (Δrok), rok complementation (Δrok + rok), complementation with rok mutant with 45 aa truncation from the N-terminus (Δrok + rok45) and complementation with rok mutant with 95 aa truncation from the N-terminus (Δrok + rok95). The interaction between Rok clusters 1 and 6 is shown in the inset for each strain. b) Interaction between Rok terminus supercluster in the wild type and the different rok mutant strains. The region shown in b) is marked in the Hi-C map of wild type strain in a).
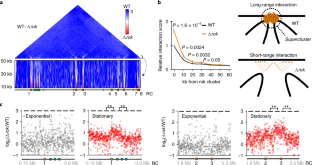
Extended Data Fig. 3 Inhibition of replication promotes Rok mediated chromosomal loop formation in exponential phase.
Normalized Hi-C contact maps of wild type strain at exponential phase after treatment with the replication inhibitor hydroxyurea (1 mg/ml). The interaction between Rok clusters 1 and 6 is shown in the inset. Hydoxyurea shows some inhibition of the SMC complex (reduced contacts in the secondary vertical diagonal showing juxtaposition of the two chromosome arms), presumably since this complex traverses from the origin to the terminus and encounters arrested replisomes.
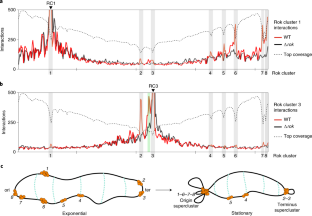
Extended Data Fig. 4 Virtual 4 C analysis of Rok clusters at the origin supercluster.
Interactions of Rok clusters 6 and 7 with the whole genome during stationary phase in wild type (wt) and Δrok strains. Rok clusters are marked using grey bars. DNA coverage of top fraction obtained from the wild type strain is shown as dotted line.
Extended Data Fig. 5 Virtual 4 C analysis of Rok clusters at the terminus supercluster.
a) Interactions of Rok cluster 2 with the whole genome during stationary phase in wild type and Δrok strains. Rok clusters are marked using grey bars. b) Interactions of a Rok binding site (located between Rok cluster 2 and 3, near yonX gene) with the whole genome during stationary phase in wild type and Δrok strains. This site was found to interact with both Rok cluster 2 and 3 (see Fig. 4b) and is recruited to the Rok terminus supercluster during stationary phase. Rok ChIP data1 (orange) at the terminus supercluster is shown below. The Rok binding site is marked using green bar. DNA coverage of top fraction obtained from the wild type strain is shown as dotted line.
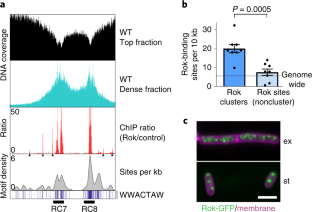
Extended Data Fig. 6 Three other Rok ChIP sites interact with the Rok origin supercluster.
a) Normalized Hi-C contact maps of wild type and Δrok strains near Rok cluster 1 at stationary phase. Rok ChIP data (red) is also shown along the genome below highlighting the other Rok binding sites as green dots. b) Difference plot shows Rok dependent interaction of three other Rok binding sites (green dots) with Rok clusters (1, 6,7 and 8) from the origin supercluster. c) Illustration shows association of Rok clusters 1,6,7 and 8 to form the origin supercluster and their interaction with the three nearby Rok-binding sites recruited to the origin supercluster at stationary phase.
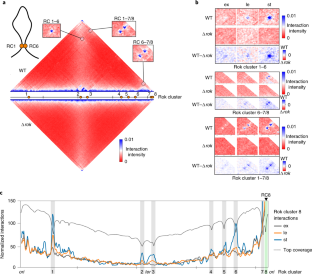
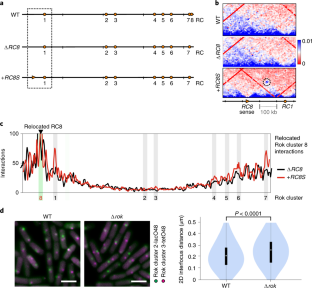
Extended Data Fig. 7 Relocated Rok cluster 8 (RC8) interacts with other Rok clusters.
a) Location of Rok clusters in the wild type and modified genomes. RC8 was deleted and complemented at an ectopic locus (amyE) within the origin supercluster in both sense (+RC8S) and reverse (+RC8R) orientation. b) Virtual 4 C analysis to determine the interactions of the amyE locus (containing the complemented RC8) with the whole genome during stationary phase in ΔRC8 and the complemented strains. Rok clusters are marked using grey bars and RC8 complementation at the amyE locus is marked using a green bar. c) Screenshot showing relative cDNA reads (RNA-seq data) mapped to RC8 in the wild type, RC8 deletion and the RC8 complementation strains (without yybN promoter). The transcription start site and the terminator around the yybN gene obtained from SubtiWiki is marked in the annotation below. d) Normalized Hi-C contact maps and difference plots of wild type and the RC8 mutant strains near the amyE complementation locus at stationary phase. The genomic regions shown is boxed in a).
Extended Data Fig. 8 SMC complex mediates Rok cluster interactions.
a) Circular representation of DNA coverage from top fraction of differential sedimentation assay (wild type coverage from Fig. 1) after smoothing. The other three Rok binding sites which were found to interact with the origin supercluster (see Extended Data Fig. 6) are marked using an asterisk. b) Normalized Hi-C contact maps of wild type (wt) and ΔscpB strains along with the difference plot at stationary growth phase in minimal media (SMM). The SMC protein forms a homodimer, and together with the kleisen protein ScpA and the kite protein ScpB it forms the SMC complex. Deletion of scpB inactivates the SMC-complex. The interaction between Rok clusters 1 and 6 is shown in the inset for both strain. The ΔscpB strain is only viable when grown in minimal medium. However, Rok cluster are also formed in wild type strain during stationary growth phase in minimal medium (c).
Extended Data Fig. 9 Role of DnaA in Rok mediated chromosomal loop formation.
a) Normalized Hi-C contact maps of wild type and dnaA deletion strains at exponential phase. b) Normalized Hi-C contact maps of wild type, dnaA deletion and rok deletion strains at stationary phase. The interaction between Rok clusters 1 and 7/8 is shown in the inset.
Extended Data Fig. 10 Topological domains boundaries are formed by interaction between Rok clusters.
a) Scalogram and DI analysis (200 Kb scale) of wild type and Δrok strains near Rok cluster 1 and 3 other Rok binding sites (green dots) recruited to the origin supercluster at stationary growth phase (see Extended Data Fig. 6). b) DI analysis (400 Kb scale) of RC8 mutant strains (see Extended Data Fig. 7) at stationary growth phase. The RC8 complementation locus is marked using a dotted line.
Supplementary information
Rights and permissions
About this article
Cite this article
Dugar, G., Hofmann, A., Heermann, D.W. et al. A chromosomal loop anchor mediates bacterial genome organization. Nat Genet 54, 194–201 (2022). https://doi.org/10.1038/s41588-021-00988-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00988-8



