Abstract
Given the critical role of SCF E3 ligases in autophagy by modulating the protein stability of various autophagic components, the activity of SCF should be tightly controlled to maintain the autophagic flux. We here showed that Numb, a multifunctional adaptor protein, increased the protein abundance of DEPTOR, which is an inhibitor of mTORC1, leading to increased autophagy flux. In vitro ubiquitination assay demonstrated that Numb inhibited SCFβ-TrCP2 mediated ubiquitination of DEPTOR. Mechanistically, Numb interrupted the interaction between β-TrCP2 and SKP1 by directly binding with SKP1. In the presence of wild type β-TrCP2, Numb overexpression inhibited DEPTOR degradation. Whereas, in the presence of the mutant β-TrCP2 which lacks the F-box domain, Numb overexpression did not affect the protein abundance of DEPTOR. In mouse model of renal fibrosis induced by unilateral ureteral obstruction, the expression of Numb was significantly increased. Consistently, the upregulation of Numb was observed in renal fibrotic lesions of chronic kidney disease patients. Specifically depleting Numb in proximal renal tubules decreased the protein abundance of DEPTOR, attenuated autophagy in fibrotic lesions and protected the kidney from development of renal fibrosis in vivo. Taken together, both in vitro and in vivo data indicated that Numb functions as a novel regulator to fine tuning the activity of SCFβ-TrCP2 in modulating autophagy.
Similar content being viewed by others
Introduction
Autophagy is an evolutionary conserved pathway which is essential to maintain cellular homeostasis through lysosome degradation [1,2,3]. Autophagy recycles needless cytoplasmic substances and organelles in basal condition, and be activated in response to intracellular and extracellular stimuli such as energy reduction, DNA damage, and cell death stress [4,5,6,7]. The dysregulated autophagy has been proved to be involved in a series of human disorders [8,9,10].
The modulating mechanisms of autophagy have been extensively investigated. Mammalian target of rapamycin complex 1 (mTORC1) has been recognized as a key negative regulator of autophagy [11,12,13], which consists of RAPTOR (Regulatory-associated protein of mTOR), DEPTOR, mTOR and GβL (Target of rapamycin complex subunit LST8). Under nutrient-rich conditions, RHEB activates mTORC1 with the assistance of Rag GTPases [14], thus, mTORC1 binds to ULK1 and maintain the inactivated state of ULK1. In low energetic state, AMPK (AMP-activated protein kinase) is activated to turn off mTORC1 through phosphorylating RAPTOR. In response to starvation, DEPTOR, a negative regulator, is elevated leading to the dissociation of mTORC1 from ULK1 and allowing its activation [15].
Increasing evidence indicate that post-translational modifications regulate the autophagy by modulating the activity, stability, and assembly of autophagic components [16,17,18]. Cullin-RING ligases (CRLs) is the largest family of E3 ubiquitin ligases in mammalian cells [19]. The SCF (SKP1 (S-phase-kinase-associated protein 1), Cullin-1, F-box protein) E3 ligases, the founding members of CRLs, consist of adaptor protein SKP1, scaffold protein cullin-1, the F-box protein, and one out of two RING (Really Interesting New Gene) family proteins RBX1(RING-box protein 1) or RBX2 (RING-box protein 2). The F-box proteins, by binding to SKP1 and cullin-1 through the F-box domain and substrates through their WD40 or leucine-rich repeats (LRRs) domains, determine the substrate specificity of the SCF complex. Several components of autophagy machinery, as well as regulators of autophagy, are subjected to SCF-mediated ubiquitination. For example, DEPTOR, a direct mTOR inhibitor, undergoes ubiquitin-mediated degradation by SCFβ-TrCP. Second, REDD1, an inhibitor of mTORC1, is subjected to CUL1β-TrCP-mediated ubiquitination and degradation with GSK-3β as a corresponding kinase for phosphorylation. Third, PDCD4, a substrate of SCFβ-TrCP [20], negatively regulates ATG5 expression and autophagy [21]. Forth, RagA inhibits mTORC1 lysosomal localization and activation relies on K63-linked ubiquitinated by SCFSKP2 [22]. These findings indicated that the activity of SCF complex must be tightly regulated during the process of autophagy, to make sure the autophagic modulators are exactly regulated. Therefore, the mechanisms that govern SCF activity should be explored thoroughly.
Numb is an evolutionary conserved multi-function protein that is firstly discovered as an cell fate determinant during the development of nervous system in Drosophila [23]. Further studies revealed that Numb is ubiquitously expressed in various tissues and functions as adaptor protein regulating both clathrin-dependent and clathrin-independent endocytic of multiple molecules including Notch [24], integrin [25], E-cadherin [26], etc. In addition, Numb interacts with a couple of E3 ligases, including Mdm2 [27, 28] and Itch [29, 30], via its N-terminal PTB domain and thus modulates their ligase activity. These findings suggest that Numb might play a regulatory role in autophagy. In this study, we provided evidence that Numb interrupts the interaction between β-TrCP2 and SKP1 by directly binding with SKP1, inhibits SCFβ-TrCP2-mediated DEPTOR protein degradation, and thus promotes autophagic flux.
Results
Numb promotes autophagosome formation
To evaluate the role of Numb in autophagy, FLAG-tagged Numb was transfected into HEK293 cells. Compared with vector transfected cells, Numb overexpression increased the protein abundance of LC3B-II at basal level, as well as in EBSS starved (Fig. 1A) and rapamycin treatment condition (Fig. 1C). As expected, the protein level of SQSTM1/p62 was decreased in Numb overexpressing cells at basal level, and in EBSS starved (Fig. 1B) and rapamycin treatment condition (Fig. 1D). Immunofluorescence staining confirmed that the number of endogenous LC3B puncta was significantly increased in Numb-overexpressing cells (Fig. 1E, F). In addition, forced expression of p72, p71, p66, or p65, which are four isoforms of mammalian Numb, showed similar effect on LC3B-II accumulation (Supplementary Fig. 1). Next, we knocked down endogenous Numb in HEK293 cells using CRISP/Cas9 technique (Numb-KO cells). Western blotting showed that Numb depletion significantly decreased the level of LC3B-II, increased SQSTM1 at basal level, and in EBSS starved and rapamycin treatment condition (Fig. 1 G–I).
A Vector or FLAG-tagged Numb was transfected into HEK293 cells for 24 h, then the cells were treated with EBSS for indicated time. Western blotting analyses (left panel) and quantitative data of LC3B protein level (right panel) are presented. B Vector or FLAG-tagged Numb was transfected into HEK293 cells for 24 h, then the cells were treated with EBSS for indicated time. Western blotting analyses (upper panel) and quantitative data of SQSTM1 protein level (lower panel) are presented. C HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, then treated with 10 nM of rapamycin for indicated time. Western blotting analyses (left panel) and quantitative data of LC3B protein level (right panel) are presented. D HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, then treated with 10 nM of rapamycin for indicated time. Western blotting analyses (left panel) and quantitative data of SQSTM1 protein level (right panel) are presented. E, F Representative images of the LC3B puncta in control or Numb overexpressing cells under EBSS or rapamycin treatment. Scale bar, 10 µm. To quantify the numbers of LC3B puncta, more than 80 cells were scored. G–I WT or Numb-KO cells were incubated with rapamycin (10 μM) or EBSS for 2 h. Western blotting analyses show the protein levels of LC3B-II and SQSTM1. Statistical analysis was performed using t-test. Data are means ± SEM of six independent experiments, ***p < 0.001.
The augmented LC3B-II accumulation in Numb-overexpressing cells might result from either enhanced autophagosome formation or decreased autophagosome degradation [31]. To address this issue, Numb-overexpressing cells were treated with BafA1 for 2 h. Surprisingly, BafA1 treatment did not affect Numb-elevated LC3B-II in growth media and EBSS treatment condition (Fig. 2A). In contrast, Wortmannin, an inhibitor of PI3K that has been widely used to stop autophagosome formation, totally abolished the increase of LC3B-II in Numb overexpressing cells in growth media and EBSS treatment condition (Fig. 2C, D). Similar result was obtained when cells were treated with another PI3K inhibitor LY294002 (Supplementary Fig 2). Furthermore, depletion of PTEN, an endogenous antagonist of PI3KC1, decreased the level of LC3B II level in empty-vector transfected cells, but not in Numb over-expression cells (Fig. 2E, F). Moreover, a GFP-RFP-LC3B reporter assay was employed to evaluate the autophagic flux. Forced expression of Numb significantly induced autophagy flux in both basal and EBSS treatment conditions, as shown by the increased number of RFP-positive only LC3B puncta (Fig. 2G–H). Collectively, these results suggest that Numb functions downstream of PI3KC1 to promote autophagosome formation.
A, B HEK293 cells were transfected with vector or FLAG-tagged Numb p72 for 24 h, then treated with EBSS with or without BafA1 (100 nM) for 2 h. Western blotting analyses show the protein levels of LC3B-II. C, D HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, and then incubated with or without EBSS and Wortmannin (10 μM) for 2 h. Western blotting analyses show the protein levels of LC3B-II. E, F HEK293 cells were co-transfected with Numb together with negative control siRNA or siPTEN for 24 h. The levels of LC3B were measured by Western blotting. G, H HEK293 cells were co-transfected with Numb and RFP-GFP-LC3B for 24 h, then incubated with or without EBSS for 3 h. Representative confocal images of LC3B puncta (G) and quantification of numbers of RFP-positive and GFP-negative LC3B-labeled autolysosomes (H) were shown. More than 50 cells were scored. Scale bar, 5 µm. Statistical analysis was performed using t-test. Data are means ± SEM of six independent experiments, **p < 0.01, ***p < 0.001, NS not significant.
Numb regulates the activity of mTORC1
Given the critical role of mTORC1 in modulating autophagy induction [32, 33], we assessed whether Numb modulates mTORC1 activity. Western blotting showed that Numb overexpression significantly decreased the level of phosphorylated S6K (p-S6K) in both nutrient-rich and EBSS treatment conditions. In addition, the activity of UNC-51-like kinase (ULK1) was further increased in Numb over-expressing cells after EBSS treatment, which is evidenced as phosphorylation at serine 757 (p-ULK1) (Fig. 3A–C). In contrast, Numb depletion increased the level of p-S6K and p-ULK1 in both growth media and EBSS treatment condition (Fig. 3D–F). In addition, we examined the effect of Numb on the activity of mTORC2 using pS473-AKT as a readout. Numb overexpression decreased the level of pS473-AKT (Fig. 3G). While, Numb depletion increased the level of pS473-AKT in both growth media and EBSS treatment condition (Fig. 3H). These data indicated that Numb overexpression inhibited the activity of mTORC1 and mTORC2.
A–C HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, and treated with EBSS for 2 h. The levels of phosphorylated S6K, total S6K, phosphorylated ULK1, total ULK1 were measured by Western blotting. D–F WT or Numb-KO cells were treated with EBSS for 2 h. The levels of phosphorylated S6K, total S6K, phosphorylated ULK1, total ULK1 were measured by Western blotting. G HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, and treated with EBSS for 2 h. The levels of phosphorylated AKT and total AKT were measured by Western blotting. H WT or Numb-KO cells were treated with EBSS for 2 h. The levels of phosphorylated AKT and total AKT were measured by Western blotting. I Vector or FLAG-tagged Numb isoform p72 was transfected into HEK293 cells for 24 h. The protein levels of p-JNK, total JNK, p-AMPK, total AMPK were measured by Western blotting. Statistical analysis was performed using t-test. Data are means ± SEM of six independent experiments, ***p < 0.001.
Endogenous JNK has been reported to be required for starvation-induced Bcl-2 phosphorylation, and regulated autophagic activity in response to nutrient limitation [34, 35]. However, the level of phosphorylated JNK (p-JNK) was not changed by Numb overexpression (Fig. 3I). In addition, Numb overexpression did not interfere with the level of phosphorylated AMPK (p-AMPK), a key cellular energy sensor and functions to trigger autophagy upon nutrient starvation (Fig. 3I). Collectively, these data suggest that Numb overexpression augmented autophagy induction at least partially by inhibiting mTORC1 activity.
Numb positively regulates DEPTOR protein stability
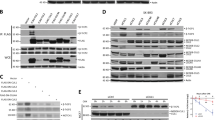
To dissect the mechanism by which Numb inhibits mTORC1 activity, we examined the expression of mTORC1 complex. As shown in Fig. 4A, B, Numb overexpression significantly increased the abundance of DEPTOR, in both basal and EBSS treatment condition, but did not cause any obvious change to the level of GβL, RAPTOR and mTOR. Conversely, the protein level of DEPTOR was decreased in Numb-KO cells in both growth media and EBSS treatment condition (Fig. 4C, D). Next, we measured the protein levels of REDD1 and TSC2, which are inhibitors of mTOR. As shown in Supplementary Fig. 3, Numb over-expression increased the protein level of REDD1, but not TSC2.
A, B HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h, and then treated with EBSS for 2 h. The levels of DEPTOR, RAPTOR, mTOR, and GβL were measured by Western blotting. Quantitative data of DEPTOR protein level (B) are presented (t-test, ***p < 0.001, n = 6). C, D WT or Numb-KO cells were treated with EBSS for 2 h. The levels of DEPTOR were measured by Western blotting. Quantitative data of DEPTOR protein level (D) are presented (t-test, ***p < 0.001, n = 6). E The mRNA level of DEPTOR in empty vector or Numb transfected cells (t-test, NS not significant, n = 5). F, G WT or Numb-KO cells were treated with cycloheximide (CHX) for indicated time period. The protein levels of DEPTOR proteins were measured by western blotting (F). Half-lives were derived from simple linear regression using Prism 8 (G). H, I HEK293 cells were transfected with vector or FLAG-tagged Numb for 24 h. Whole cells lysates were subjected to immunoprecipitation (IP) using goat IgG or anti- mTOR antibody and analyzed by immunoblotting using indicated antibodies. The ratio of band density of immunoprecipitated DEPTOR relative to immunoprecipitated mTOR was presented (t-test, **p < 0.01, n = 5) (I). J–L HEK293 cells were co-transfected with Numb together with negative control siRNA or siDEPTOR for 24 h, and then treated with or without EBSS for 2 h. The levels of phosphorylated S6K, total S6K, phosphorylated ULK1 and total ULK1 were measured by Western blotting. Representative western blots (J) and quantitative data of p-ULK1/Total ULK1 (K) and p-S6K/ Total S6K (L) are presented (t-test, ***p < 0.001, n = 6).
To determine at which regulatory level Numb controls DEPTOR expression, we conducted real time PCR. In Numb overexpressing cells, the mRNA level of DEPTOR remained almost unchanged (Fig. 4E). Furthermore, when protein synthesis was blocked by cyclohexamine (CHX), Numb depletion significantly shortened the half-life of endogenous DEPTOR protein (Fig. 4F, G). Furthermore, we conducted co-immunoprecipitation (Co-IP) assay using anti-mTOR antibody. As shown in Fig. 4H–I, Numb overexpression led to an increased association of endogenous DEPTOR with mTOR, but did not influence the abundance of RAPTOR or GβL to mTOR. Moreover, the reduced levels of p-ULK1 and p-S6K by Numb overexpression were rescued when endogenous DEPTOR was silenced by siRNA (Fig. 4J–L). These data indicate that Numb acts as a negative regulator of mTORC1 activity via regulating DEPTOR protein degradation.
Numb interferes with SCFβ-TrCP2-mediated DEPTOR ubiquitination
Since DEPTOR is the substrate of SCFβ-TrCP2 for ubiquitin-mediated degradation [36], we next asked whether Numb inhibits the E3 ligase activity of SCFβ-TrCP2. To assess this possibility, we treated cells with MLN4924, an inhibitor of the NEDD8-activating enzyme [37, 38], to inhibit the activity of CUL1. As shown in Fig. 5A, the decreased protein level of DEPTOR in Numb-KO cells was blocked by MLN4924. Furthermore, silencing endogenous CUL1 or β-TRCP2 rescued the protein abundance of DEPTOR in Numb-KO cells (Fig. 5B, C). To conduct in vitro ubiquitination assay, Flag-CUL1 and His-tagged β-TrCP2 was transfected into HEK293 cells, respectively. The complex co-immunoprecipitated with Flag-CUL1 or His-β-TrCP2 was used as E3 in in vitro ubiquitination assay, respectively. As shown in Fig. 5D, E, the existence of immunopurified Numb significantly inhibited CUL1-SKP1-β-TrCP2 mediated ubiquitination of DEPTOR.
A WT and Numb KO cells were treated with or without MLN4924 for 12 h. The protein levels of DEPTOR and Numb were analyzed by western blotting. Representative western blots (upper panel) and quantitative data of DEPTOR level (lower panel) are presented (t-test, ***p < 0.001, n = 6). B WT and Numb KO cells were transfected with negative-control (NC) siRNA or CUL1 siRNA. The protein level of DEPTOR and CUL1 were measured by Western blotting. Representative western blots (upper panel) and quantitative data of DEPTOR level (lower panel) are presented (t-test, ***p < 0.001, NS not significant, n = 6). C WT and Numb KO cells were transfected with negative control siRNA or β-TrCP2 siRNA, respectively. The protein level of DEPTOR and β-TrCP2 were measured by Western blotting. Representative western blots (upper panel) and quantitative data of DEPTOR level (lower panel) are presented (t-test, ***p < 0.001, NS not significant, n = 6). D In vitro DEPTOR ubiquitination assay using immunopurified FLAG-tag CUL1/RBX1 and Myc-DEPTOR from HEK293 cells together with recombinant His-Ubiquitin. Reactions were performed in the presence or absence of E1 (UBA1) /E2 (UbcH5c) or CUL1 to verify ubiquitination reaction specificity. DEPTOR ubiquitination were evaluated upon IP by immunoblotting using anti-His antibody. E In vitro DEPTOR ubiquitination assay using immunopurified His-tag β-TrCP2 and Myc-DEPTOR from HEK293 cells together with recombinant Flag-Ubiquitin. Immunopurified SKP1Flag-β-TrCP2 complex was used as the E3, and reactions were performed in the presence or absence of E1 (UBA1)/E2 (UbcH5c) or E3. DEPTOR ubiquitination were evaluated upon IP by immunoblotting using anti-Flag antibody.
Numb directly interacts with SKP1 leading to the dissociation of β-TrCP2 from SKP1
To elucidate the mechanism by which Numb regulates the activity of SCF complex, we conducted GST-PTB pull down using PTB domain of Numb as the bait. The protein complex binding to Numb-PTB was identified by mass spectrometry. As shown in Table 1, CUL4B, DNA damage-binding protein 1 (DDB1), S-phase kinase-associated protein 1 (SKP1), Elongin-B (ELOB), COP9 signalosome complex subunit 1 (CSN1), and COP9 signalosome complex subunit 5 (CSN5) were identified. Since DEPTOR is the substrate of CUL1, we conducted GST- PTB pull down assay to confirm the interaction of Numb with CUL1 and SKP1. As shown in Fig. 6A, Numb PTB domain was able to pull down endogenous SKP1, exogenous His-tagged CUL1. In addition, in Flag-tagged full length Numb, but not a mutant Numb lacking PTB domain (Numb-ΔPTB), transfected cells, endogenous SKP1 and CUL1 were co-precipitated with Numb using anti-FLAG antibody (Fig. 6B), indicating that the PTB domain mediates the interaction of Numb with SKP1 and CUL1. Consistently, the protein level of DEPTOR was not elevated in Numb-ΔPTB overexpressing cells (Supplementary Fig. 4). Besides DEPTOR, the protein levels of Cyclin E and p27, which are substrates of SKP1-CUL1, were increased in Numb-overexpressing cells, but decreased in Numb-KO cells (Supplementary Fig. 5).
A Whole cell lysates from His-CUL1 transfected HEK293 cells were incubated with purified GST-Numb PTB or GST, respectively. Bound protein was detected by Western blotting using anti-β-TrCP2, anti-SKP1, or anti-His antibody. B HEK293 cells were transfected FLAG-Numb (full length) or PTB domain truncated (ΔPTB) for 24 h. Total cell extracts were subjected to IP using indicated antibody, and analyzed by Western blotting using anti-CUL1, anti-SKP1 or anti-Flag antibody. C Numb binds to SKP1 in vitro: His-tag Numb were expressed and purified from BL21 E. coli strain, then incubated with bead-conjugated GST-SKP1 or GST peptide, bound proteins were detected by IB using anti-His antibody. D Numb binds to SKP1 in vitro: His-tag Numb were expressed and purified from BL21 E. coli strain, then incubated with bead-conjugated GST-SKP1 full length, GST-SKP1-ΔN, GST-SKP1-C or GST alone peptide, followed by IB using anti-His Ab detected the bead-conjugated proteins. E WT or Numb KO cells were transfected with His-SKP1 for 24 h. Total cell extracts were subjected to IP using either an anti-His or anti-IgG antibody, and analyzed by IB using anti-His and anti-β-TrCP2 antibody. The ratio of band density of immunoprecipitated β-TrCP2 relative to immunoprecipitated His- SKP1 was presented in lower panel (t-test, ***p < 0.001, n = 5). F HEK293 cells were co-transfected His-SKP1 with empty vector, FLAG-Numb (full length) or PTB domain truncated (ΔPTB) for 24 h, respectively. Total cell extracts were subjected to IP using either an anti-His or anti-IgG antibody and analyzed by IB. G HEK293 cells were transfected empty-vector or FLAG-Numb, or Numb-ΔPTB for 24 h, and then treated with EBSS for indicated time period. Western blotting analyses showed the protein levels of LC3B. H HEK293 cells were transfected β-TrCP2 siRNA for 24 h, and then co-tranfected WT β-TrCP2 or mutant β-TrCP2 which lacks the F-box domain with empty-vector or FLAG-Numb for another 24 h. The amount of WT β-TrCP2 or mutant β-TrCP2 plasmid were adjusted to make sure the equal expression of exogenous β-TrCP2 protein between vector and Numb over-expression cells. The protein level of DEPTOR was measured by Western blotting. Representative western blots and quantitative data of DEPTOR level are presented (t-test, ***p < 0.001, NS not significant, n = 6). I HEK293 cells were co-transfected His-SKP1 with vector or FLAG-tagged Numb for 24 h, and then treated with EBSS for indicated time period. Total cell extracts were subjected to IP using either an anti-His or anti-IgG antibody, and analyzed by immunoblotting using indicated antibody.
Next, to determine whether Numb interacts with SKP1 directly, the association of bacterially produced GST-SKP1 with His-Numb was analyzed in vitro. GST-SKP1, but not GST alone, bound to His-tagged Numb (Fig. 6C). Next, we generated a series of different parts of SKP1 truncations, including SKP1 lacking the N-terminal (SKP1-ΔN) and SKP1 C-terminal only (SKP1-C). In vitro binding assay demonstrated that both SKP1-ΔN and SKP1-C can bind to Numb (Fig. 6D). Collectively, these data indicate that Numb PTB domain directly interacts with the C-terminal of SKP1.
Since C-terminal of SKP1 mediates the binding with F-box proteins [39], we wonder whether Numb interferes with the interaction between β-TrCP and SKP1. Most recent study showed that β-TrCP2 preferentially degrades DEPTOR [36]. We thus specifically analyzed the binding of β-TrCP2 with SKP1. Interestingly, the protein level of β-TrCP2 was reduced in Numb-KO cells. While, the protein level of β-TrCP1 was also reduced but not as much as that of β-TrCP2 (Supplementary Fig. 6). Co-IP assay revealed that the amounts of endogenous β-TrCP2 co-precipitated with SKP1 were increased although the total amount of β-TrCP2 was reduced (Fig. 6E). In Numb-overexpressing cells, the amounts of β-TrCP2 co-precipitated with SKP1 were reduced. However, the association of β-TrCP2 with SKP1 was not affected in Numb-ΔPTB-overexpressing cells (Fig. 6F). Then we determined the role of PTB domain in regulating autophagy. HEK293 cells were transfected with empty-vector, full-length Numb or Numb-ΔPTB, respectively. The protein level of LC3B were measured by western blotting in both basal and EBSS treatment condition. As shown in Fig. 6G, full-length Numb, but not Numb-ΔPTB, increased the LC3B II level, suggesting PTB domain mediates the modulating effect of Numb on autophagy.
To clarify whether the interaction between β-TrCP2 and SKP1 is critical for Numb-mediated DEPTOR degradation, β-TrCP2 was knocked down by siRNA to eliminate the interference of endogenous β-TrCP2, and then wild type β-TrCP2 was co-transfected with either empty vector or Numb into cells, respectively. Compared with empty vector, Numb overexpression increased DEPTOR protein (Fig. 6H). Next, we generated a mutant β-TrCP2 which lacks the F-box domain to disrupt the interaction between β-TrCP2 and SKP1, and then the mutant β-TrCP2 was co-transfected with either empty vector or Numb into cells depleted endogenous β-TrCP2. Compared with empty vector, Numb overexpression did not change the protein level of DEPTOR (Fig. 6H), indicating that the effect of Numb on DEPTOR degradation at least partially dependent on the β-TrCP2-SKP1 interaction.
Prompted by the above findings, we next examined whether Numb interferes with the association of DEPTOR with SKP1 during the process of autophagy. Western blotting revealed that the amount of endogenous DEPTOR was significantly increased after 2 h EBSS treatment, and decreased slightly after 12 h. Co-IP assay revealed that the association of DEPTOR with SKP1 was decreased after 2 h EBSS treatment, but partially reversed at 12 h although the total amount of DEPTOR protein was decreased (Fig. 6I). However, Numb overexpression disrupted the dynamic association of DEPTOR/β-TrCP2 with SKP1 during the process of autophagy. Noteworthy, the amount of endogenous β-TrCP2 was decreased after 12 h EBSS treatment together with decreased endogenous Numb. In addition, β-TrCP2 was significantly increased in Numb overexpressed cells. These data indicated that Numb also implicates in the degradation of β-TrCP2 during the process of autophagy.
Numb regulates the stability of DEPTOR protein and autophagy in vivo
We previously reported that Numb is upregulated in mouse model of TIF induced by unilateral ureteral obstruction (UUO). We further investigated the expression of Numb in Nakagawa CKD Kidney Dataset (Nephroseq database [40]), which includes renal biopsies from 53 chronic kidney disease (CKD) patients and eight controls. Compared with healthy controls, the mRNA level of Numb was significantly upregulated in renal biopsies of CKD patients (Fig. 7A). These data demonstrated the upregulation of renal Numb in both mouse and human CKD.
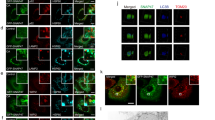
A Increased Numb expression in chronic kidney disease from Nakagawa CKD Kidney Dataset in the Nephroseq platform (Mann–Whitney U-test, p = 0.0019, fold change = 2.23). B Immunohistochemical staining of Numb and β-TrCP2/ DEPTOR in sequential sections of UUO kidneys. Co-localization of Numb with β-TrCP2/ DEPTOR in the same tubules were marked with stars. Bar = 50 μm. C Immunoblot analyses of proteins in the sham-operated and UUO kidneys in PT-Nb-WT and PT-Nb-KO mice (left panel). Quantitative data of indicated proteins normalized to GAPDH (right panel) are presented (two-way ANOVA, **p < 0.01, ***p < 0.001, n = 6). D–G Immunoblot analyses of proteins levels of Fibronectin, COL I and α-SMA in the UUO kidneys in PT-Nb-WT and PT-Nb-KO mice. Representative western blots (D) and quantitative data of Fibronectin (E), COL I (F) and α-SMA (G) levels was presented (t-test, ***p < 0.001, n = 6).
Next, we explored whether Numb regulates the stability of DEPTOR protein and autophagy in vivo. Immunohistochemical staining showed that Numb, DEPTOR and β-TrCP2 were localized in the same segment of renal tubules in UUO kidney (Fig. 7B). Furthermore, we employed a conditional knockout mouse model with Numb specifically deleted from renal proximal tubules (PT-Numb-KO) which has been generated by our lab previously [41], and subjected to UUO. As shown in Fig. 7C, the protein levels of DEPTOR and β-TrCP2 were significantly upregulated in fibrotic lesions induced by UUO together with upregulated Numb expression. Compared with control mice, the protein levels of DEPTOR and β-TrCP2 were remarkably decreased in the obstructed kidneys of PT-Numb-KO mice. Moreover, Numb depletion dramatically decreased LC3B-II protein, and blocked the degradation of SQSTM1/p62. Extracellular matrix proteins like Fibronectin, collagen I (COL I) and α-smooth muscle actin (α-SMA) expression was also decreased in Numb knock-out kidney as expected (Fig. 7D–G). These data suggest that Numb overexpression contributes to the increased abundance of DEPTOR protein and persistently activated autophagy flux in the progression of fibrosis in kidney.
Discussion
In the current study, we demonstrated that Numb functions as a positive regulator of autophagy initiation. This conclusion is supported by the following findings: first, Numb overexpression significantly increased the conversion of LC3B-I to LC3B-II and p62 degradation, two markers to measure autophagy flux, at basal level and in EBSS starved and rapamycin treatment condition. Second, Numb inhibits mTORC1, preferentially preventing DEPTOR protein degradation. Third, specific deletion Numb from renal proximal tubular cells attenuated UUO-induced autophagy in fibrotic renal tissues in vivo. Previous study reported that Numb depletion impaired autophagic flux by interfering with autophagic degradation [42]. Our finding did not exclude the involvement of Numb in autophagic degradation. One explanation for this discrepancy might be the efficiency of Numb depletion. In the current study, Numb depletion was achieved by CRISR-CAS9 knock out system. The high efficiency of CRISR-CAS9 system might result in the effect of Numb depletion on autophagic initiation more dramatic. In support of this explanation, it has been shown that clathrin is critical in both autophagosome formation and autophagic lysosome reformation. However, a highly efficient knocking out technique is required to observe the effect of clathrin on autophagosome formation, while moderate depletion reveals the requirement of clathrin in autophagic lysosome reformation [43, 44].
Post-translational modifications and the diversity of the receptor subunits are critical for SCF complex in recruiting substrates for ubiquitination. However, the E3 ligase activity is regulated mainly at the level of complex constitution. For example, CSN-5 and CUL-associated and neddylation-dissociated-1 (CAND-1) limit the SCF E3 ligase activity through disassembling the complex [45]. Our results suggest that Numb is a new negative regulator of SCFβ-TrCP2 activity by interfering with complex assembly. This conclusion is supported by the following findings: first, GST pull down and Co-IP assay revealed that Numb is in the complex of CUL1 and SKP1, but not β-TrCP2. Second, in vitro binding assay revealed that the C-terminal part of SKP1, which is critical in binding with F-box proteins, directly interacted with the PTB domain of Numb. Numb overexpression abolished the SKP1-β-TrCP2 interaction. Similarly, previous studies reported that CENP-W incorporates into the SCF complex as a substitute for SKP-1, and interacts with CUL-1 and β-TrCP1 through highly overlapped binding sites with SKP-1 [46]. Unlike CENP-W, Numb competes the binding of β-TrCP to SKP1, but not the binding of SKP1 to CUL1. Secondly, Numb-induced dissociation of β-TrCP2 from SCF complex increased the protein stability of β-TrCP2, but not promoting its degradation. Third, Numb is mainly localized in the cytosol, while CENP-W is in the nucleus. Our finding further emphasizes the complexity of the modulation of SCF function.
Noteworthy, F-box proteins share the common binding site with SKP1. Therefore, Numb might influence the protein abundance of various SCF targets. Indeed, Numb overexpression increased the protein abundance of Cyclin E (a substrate of Fbw7 [47]) and p27 (a substrate of Skp2 [48]). Consistently, previous studies have shown that Numb regulates the protein abundance of p53 (a substrate of FBXW7; [47]), Gli1(a substrate of FBXL17; [49]) and Notch1(a substrate of FBXW7; [47]).
In the current study, we demonstrated that the C-terminal part of SKP1 directly interacts with the PTB domain of Numb. However, the underlying mechanism regulating Numb and SKP1 interaction remains unexplored. Phosphorylation has been reported to modulate the physiological and pathological function of Numb. For example, phosphorylation of Numb by αPKC or PLK1 impaired Numb/p53 pathway [50]. Whether Numb phosphorylation modulates Numb-SKP1 interaction remains further investigation.
Most recent study showed that β-TrCP2 itself is also the physiological substrates of SCF E3 ligase. Consistent with previous finding, we found that β-TrCP2 protein was decreased in Numb KO cells, but elevated in Numb overexpressed cells. However, in Numb KO cells, decreasing β-TrCP2 was not accompanied with DEPTOR accumulation. This discrepancy might be caused by the following reasons: First, co-IP assay revealed that, in Numb KO cells, the binding of β-TrCP2 to SKP1 was increased although the protein level of β-TrCP2 was decreased (Fig. 6E), suggesting that limited amount of β-TrCP2 is sufficient to recruit DEPTOR to SKP1-CULI complex. Second, both β-TrCP1 and β-TrCP2 function as the substrate adaptor of DEPTOR. In the absence of β-TrCP2, DEPTOR could be recognized by β-TrCP1. In supporting this hypothesis, we found that, in Numb KO cells, the protein level of β-TrCP1 was reduced not as much as that of β-TrCP2 (Fig Supplementary Fig. 6). Previous studies showed that β-TrCP1/2 homodimerization promotes their E3 activity against the substrates and that heterodimerization mediates their own degradation [36]. In Numb KO cells, the lower amount of β-TrCP2 might facilitate the homodimerization of β-TrCP1 for DEPTOR degradation.
We previously reported that the expression of Numb was upregulated in the fibrotic lesions and promoted renal interstitial fibrosis via causing cell cycle arrest of proximal tubular cells. In the current study, we provided further evidence that the expression of Numb was significantly upregulated in renal biopsies of CKD patients (Fig. 7A). According to the literatures, the key components of Sonic Hedgedog signaling (Gli1) [49], Notch signaling (Notch1) [51], and Wnt signaling (β-catenin) [52] are substrates of SCF complex. Therefore, Numb might also contribute to the constitutive activation of these signaling pathways in the progression of renal fibrosis. Besides the upregulation of Numb in fibrotic kidney, the downregulation of Numb has been observed in various tumor tissues [53,54,55]. On the contrary, SKP1 with oncogenic SCF assembly components promotes development breast, colon, prostate, lung and gastric cancers [56]. Our data suggest that Numb might exert its tumor suppressor function through modulating the function of SCF complex.
In summary, our data revealed that Numb is a novel regulator of SCFβ-TrCP2. Numb directly interacts with SKP1, and thus interferes with the SKP1-β-TrCP2 interaction leading to the accumulation of DEPTOR and enhanced autophagic flux. Specifically depleting Numb from renal tubules decreased DEPTOR protein, attenuated autophagy and renal fibrosis. These novel findings will provide valuable therapeutic insights for the treatment of chronic kidney disease.
Materials and methods
Plasmid constructs and siRNAs
The full-length cDNA of Numb, CUL1, SKP1, β-TrCP2 and RBX1 were cloned into CMV promoter Vector (Beyotime, Shanghai, China), or pGEX4T-2 (General Electric Company, Boston, USA), or pET-28a (+) (Novagen, Massachusetts, USA). Ptf-GFP-RFP-LC3B was purchased from Addgene (#21074). Oligonucleotide siRNA duplex was synthesized by RiboBio, Guangzhou, China. RNAi oligonucleotides sequences are as follows: DEPTOR: CGGCACCTTCCCATTGGATAATGAA, CUL1#1: AGGAACAACGAAGAGTTCAGGTTTA, CUL1#2: GCCCTTCCAGCAGTCTTGTACATTT. β-TrCP2: GCCCATGTTGCAGCGGGACTTTATT.
Cell culture and transfection
HEK293 cells were grown in DMEM (Thermofisher, Guangzhou, China) with 10% FBS (Biological Industries, Israel). Earl’s balanced salt solution (Thermofisher, Guangzhou, China) was used as the starvation medium. Transfection with siRNA or plasmid were using jetPRIME reagent (Polyplus, Illkirch, France), following the protocols in the instruction.
Generation of Numb-knockout cell lines
HEK293 cells were co-transfected with two lentiCRISPR v2 plasmids [57] containing different sgRNA targeting hNumb gene. sgRNA#1: GTGGCGCTTGAGTTGGTCGG, sgRNA#2: GATGAAGAAGGCGTTCGCAC. Three days after transfection, cells were diluted and seeded into 48-well plates. Seven days later, single cell clones were selected and transferred to 12-well plates. After another subculture, whole cell lysates were collected and the loss of Numb was confirmed via immunoblot. Three Numb KO colonies were selected and frozen.
Immunofluorescence staining
Immunofluorescence staining was performed using an established procedure [58, 59]. Cells were fixed with 4% (vol/vol) paraformaldehyde and incubated with primary antibodies for overnight at 4 °C, followed by incubation with Alexa Fluor secondary antibodies (Abcam, Shanghai, China). Cover slips were mounted on DAPI (Zsbio, Beijing, China) medium. Images were taken with a FV1000 scanning confocal microscope (Olympus, Tokyo, Japan). Primary antibodies were as follows: Anti-LC3B (SC-271625, SANTA Cruz, Texas, USA). Donkey anti-Mouse IgG H&L (Alexa Fluor® 594, ab150108, Abcam, Shanghai, China).
Immunoprecipitation assays and western blot analysis
Cells were lysed in IP buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40, and protease inhibitor mixture) at 4 °C for 30 min. For immunoprecipitating the mTOR complex, cells were lysed in CHAPS buffer (50 mM Tris, pH7.5, 20 mM CHAPS, 120 mM NaCl, 10% Glycerol, and protease inhibitor mixture). After incubating with indicated antibodies for 2 h on ice, the immunocomplexes were recovered using ProteinA/G Sepharose (Beyotime, Shanghai, China), resolved in SDS buffer and then loaded on SDS-PAGE for western blotting. The antibodies were used as follows: Anti-LC3B (SC-271625), and mTOR (SC-1524) were from Santa Cruz (Texas, USA). Anti -COL I (AB0325) was from Boster (Wuhan, China). Anti -FLAG(M185), HA (M180) were from MBL (Tokyo, Japan); anti-HIS (66005), DEPTOR (20985), RAPTOR (20984), GAPDH (60004), β-TrCP2(13149-1-AP) were from Proteintech (Wuhan, China). Anti-β-TrCP2 (ab137835) was from Abcam (Shanghai, China); Anti-Numb (2756 S), ser757-ULK1 (6888 S), ULK1 (D8H5), T389-S6K1 (9234 S), S6K1 (9202), DEPTOR (11816 S), GβL (3274), p-AMPK (2535), AMPK (5832), p-JNK (9255), JNK (9252), CUL1 (4995 S), β-TrCP1(4394) were from Cell Signaling (Danvers, USA). Anti-fibronectin (F3648) and anti-α-SMA (A5228) were from Millipore (Burlington, USA).
Recombinant protein purification and GST pull down assay
Recombinant protein purification and GST pull down assay was conducted as described previously [60]. Briefly, Escherichia coli (BL21) were transformed with pGEX-4T-2 or pGEX-Numb PTB domain. The GST fusion proteins were purified according to the manufacturer’s instruction (Beyotime, Shanghai, China). The GST-bound beads was then washed with GST pull-down buffer (50 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 1%NP-40, 1 mM DTT, 10 mM MgCl2, pH 8.0) and then mixed with cell lysates at 4 °C for 6 h. For western blot, the conjugated proteins were boiled with Laemmli buffer. For LS-MS/MS analysis, the beads were eluted with SDT buffer (4%SDS, 100 mM Tris/HCl, 1 mM DTT, pH 7.6).
Liquid chromatography-tandem mass spectrometry
LC/MS analysis was conducted by Applied Protein Technology (Shanghai, China). After repeated ultra-filtrating with UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0), iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues. The protein suspensions were digested with trypsin (Promega, Wisconsin, USA) and desalted on C18 Cartridges (Empore SPE Cartridges C18 (standard density), Sigma), concentrated by vacuum centrifugation and reconstituted in 0.1% (v/v) formic acid. The peptide mixture was loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, nanoViper C18) connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% Formic acid). MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Thermo Scientific). MS spectra were searched using MASCOT engine (Matrix Science, London, UK; version 2.4) against the UniProtKB database.
In vitro ubiquitination assay
In vitro ubiquitination assay was performed as previously described [61]. Briefly, HEK293 cells were transfected with His- β-TrCP2, FLAG-CUL1, FLAG-RBX1 and myc-DEPTOR, respectively. HA-Numb was overexpressed by infecting cells with adenovirus as described previously [41]. Twenty-four hours after transfection/infection, cells were lysed in buffer (20 mM HEPES, 2 mM EGTA, 1% NP-40 and 10% glycerol, PH 7.4, 1% protease inhibitors) and immunopurified using anti-FLAG, anti-HA, anti-His or anti-Myc antibodies. The assays were performed in a 20 ml reaction volume containing indicated immunopurified proteins and the components as follows: UBA1 (0.2 mg, Sinobiological, Beijing, China), UbcH5c (0.5 mg, Darmstadt, Germany), and His-ubiquitin or FLAG-ubiquitin (100 mM, Biochem, UK). The reaction was performed at 30 °C for 4 h, follow by immunoblotting with anti-His antibody.
Real-Time PCR
Total RNA was extracted using TRIzol reagent (Takara, Kyoto, Japan) [62]. Real-time RT-PCR was performed on an ABI PRISM7500 Fast system (Applied Biosystems, Foster City, CA). The primer pairs used were as follow: GAPDH: (forward): GACAGTCAGCCGCATCTTCT and (reverse) ACCAAATCCGTTGACTCCGA, DEPTOR: (forward): GAACAGCTACGGCTCAGGCT and (reverse) GCGACAAAACAGTTTGGGTAGG.
Animals
Numbflox/floxNumblikeflox/flox mice (C57BL/6 J background) were purchased from the Jackson Laboratory (Stock No. 005384). PEPCK-Cre mice were kindly provided by Dr. Volker Haase (Vanderbilt University School of Medicine, Nashville, TN). Numbflox/floxNumblikeflox/flox mice were crossed with PEPCK-Cre mice to generate the Proximal tubule-specific Numb/Numblike−/− mice. The protocol of mouse breeding and genotyping was as previously described [59]. Mice were housed in the Nanfang Hospital Animal Center. All animal experiments were performed with approval of Ethics Committee for Animal Experiments of the Southern Medical University.
For the UUO model, the left ureter was ligated using 4-0 nylon surgical sutures at the level of the lower pole of kidney, and sham-operated mice were used as controls. Mice were killed at day 7 after surgery. The kidneys were harvested for further studies.
Immunohistochemical staining
Immunohistochemical staining was performed on 4μm kidney sections. Every two sequential sections were chosen to perform staining in order to identify the same tubules. Sections were performed antigen repairing by microwave heating, then incubated with the primary antibody for 8 h at 4 °C, followed by incubating with HRP conjugated secondary antibodies for 60 min. Images were taken using an BX51 microscope (Olympus, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using Prism software (GraphPad 8). Data were presented as means ± SEM from five or six independent experiments. Comparison between two groups was using Student’s t test or Mann–Whitney U-test, as indicated. Comparison between more than two groups was using one-way or two-way ANOVA, as indicated. Significance was defined with p values. p values < 0.05 were considered significant.
Data availability
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alvers AL, Fishwick LK, Wood MS, Hu D, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–69.
Hazari Y, Bravo-San Pedro JM, Hetz C, Galluzzi L, Kroemer G. Autophagy in hepatic adaptation to stress. J Hepatol. 2020;72:183–96.
Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861.
Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–88.
Liu J, Liao G, Tu H, Huang Y, Peng T, Xu Y, et al. A protective role of autophagy in Pb-induced developmental neurotoxicity in zebrafish. Chemosphere 2019;235:1050–8.
Rabinowitz JD, White E. Autophagy and Metabolism. Science 2010;330:1344–8.
Singh K, Matsuyama S, Drazba JA, Almasan A. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy 2012;8:236–51.
Choi A, Ryter S, Levine B, Levine B. Mechanisms of disease: autophagy in human health and disease. N. Engl J Med. 2013;368:651–62.
Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword [Review]. Science 2004;306:990–5.
Wang Q, Li X, Wang Q, Xie J, Xie C, Fu X. Heat shock pretreatment improves mesenchymal stem cell viability by heat shock proteins and autophagy to prevent cisplatin-induced granulosa cell apoptosis. Stem Cell Res Ther. 2019;10:348.
Cai C, Min S, Yan B, Liu W, Yang X, Li L, et al. MiR-27a promotes the autophagy and apoptosis of IL-1beta treated-articular chondrocytes in osteoarthritis through PI3K/AKT/mTOR signaling. Aging (Albany NY). 2019;11:6371–84.
Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95.
Li T, Weng J, Zhang Y, Liang K, Fu G, Li Y, et al. mTOR direct crosstalk with STAT5 promotes de novo lipid synthesis and induces hepatocellular carcinoma. Cell Death Dis. 2019;10:619.
Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–55.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–U71.
Guo B, Liang Q, Li L, Hu Z, Wu F, Zhang P, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol. 2014;16:1215.
Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. Embo Rep. 2015;16:1071–83.
Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015;11:28–45.
Cui D, Xiong X, Zhao Y. Cullin-RING ligases in regulation of autophagy. Cell Div. 2016;11:8.
Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–60.
Wang L, Ye N, Lian X, Peng F, Zhang H, Gong H. MiR-208a-3p aggravates autophagy through the PDCD4-ATG5 pathway in Ang II-induced H9c2 cardiomyoblasts. Biomed Pharmacother. 2018;98:1–8.
Jin G, Lee SW, Zhang X, Cai Z, Gao Y, Chou PC, et al. Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent Amino-Acid-Dependent mTORC1 Hyperactivation by Recruiting GATOR1. Mol Cell. 2015;58:989–1000.
Schmit TL, Setaluri V, Spiegelman VS, Ahmad N. Abstract 1064: numb, a progenitor cell fate determinant and intrinsic inhibitor of Notch signaling, is cell cycle regulated and required for proper mitotic entry and progression in melanoma cells. Cancer Res. 2011;70:1064–1064.
Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of numb and notch. Neuron 1996;17:27–41.
Calderwood DA, Fujioka Y, de Pereda JM, Garcia-Alvarez B, Nakamoto T, Margolis B, et al. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci USA. 2003;100:2272–7.
Sato K, Watanabe T, Wang SJ, Kakeno M, Matsuzawa K, Matsui T, et al. Numb controls E-cadherin endocytosis through p120 catenin with aPKC. Mol Biol Cell. 2011;22:3103–19.
Folberg-Blum A, Sapir A, Shilo BZ, Oren M. Overexpression of mouse Mdm2 induces developmental phenotypes in Drosophila. Oncogene 2002;21:2413–7.
Kim H, Ronai ZEA. Rewired Notch/p53 by Numb’ing Mdm2. J Cell Biol. 2018;217:jcb.201712007.
Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–23.
Marcotullio LD, Greco A, Mazzà D, Canettieri G, Gulino A. Numb activates the E3 ligase ITCH to control Gli1 function through a novel degradation signal. Oncogene 2011;30:65–76.
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222.
Cavadini S, Fischer ES, Bunker RD, Potenza A, Lingaraju GM, Goldie KN, et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 2016;531:598–603.
Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through?AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–16.
Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88.
Zhu Z, Huang Y, Lv L, Tao Y, Shao M, Zhao C, et al. Acute ethanol exposure-induced autophagy-mediated cardiac injury via activation of the ROS-JNK-Bcl-2 pathway. J Cell Physiol. 2018;233:924–35.
Cui D, Dai X, Shu J, Ma Y, Wei D, Xiong X, et al. The cross talk of two family members of β-TrCP in the regulation of cell autophagy and growth. Cell Death Differ. 2020;27:1119–33.
Roberts JZ, Holohan C, Sessler T, Fox J, Crawford N, Riley JS, et al. The SCF(Skp2) ubiquitin ligase complex modulates TRAIL-R2-induced apoptosis by regulating FLIP(L). Cell Death Differ. Cell Death Differ. 2020;27:2726–41.
Toth JI, Yang L, Dahl R, Petroski MD. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 2012;1:309–16.
Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 2000;408:381–6.
Nakagawa S, Nishihara K, Miyata H, Shinke H, Tomita E, Kajiwara M, et al. Molecular markers of tubulointerstitial fibrosis and tubular cell damage in patients with chronic kidney disease. PLoS One. 2015;10:e0136994.
Zhu F, Liu W, Li T, Wan J, Tian J, Zhou Z, et al. Numb contributes to renal fibrosis by promoting tubular epithelial cell cycle arrest at G2/M. Oncotarget 2016;7:25604–19.
Sun H, Liu Y, Zhang L, Shao X, Liu K, Ding Z, et al. Numb positively regulates autophagic flux via regulating lysosomal function. Biochem Biophys Res Commun. 2017;491:780–6.
Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol. 2012;14:924–34.
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57.
Helmstaedt K, Schwier EU, Christmann M, Nahlik K, Westermann M, Harting R, et al. Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site. Mol Biol Cell. 2011;22:153–64.
Cheon Y, Lee S. CENP-W inhibits CDC25A degradation by destabilizing the SCF(β-TrCP-1) complex at G(2)/M. FASEB J. 2018;32:6051–65.
Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115.
Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, et al. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004;6:661–72.
Raducu M, Fung E, Serres S, Infante P, Barberis A, Fischer R, et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. Embo j. 2016;35:1400–16.
Siddique HR, Feldman DE, Chen CL, Punj V, Tokumitsu H, Machida K. NUMB phosphorylation destabilizes p53 and promotes self-renewal of tumor-initiating cells by a NANOG-dependent mechanism in liver cancer. Hepatology 2015;62:1466–79.
Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 2011;471:104–9.
Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–56.
Cheng C, Huang Z, Zhou R, An H, Cao G, Ye J, et al. Numb negatively regulates the epithelial-to-mesenchymal transition in colorectal cancer through the Wnt signaling pathway. Am J Physiol Gastrointest Liver Physiol. 2020;318:G841–g53.
Liang J, Han B, Zhang Y, Yue Q. Numb inhibits cell proliferation, invasion, and epithelial-mesenchymal transition through PAK1/β-catenin signaling pathway in ovarian cancer. Onco Targets Ther. 2019;12:3223–33.
Colaluca IN, Basile A, Freiburger L, D’Uva V, Disalvatore D, Vecchi M, et al. A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer. J Cell Biol. 2018;217:745–62.
Liu J, Peng Y, Zhang J, Long J, Liu J, Wei W, Targeting SCF. E3 Ligases for Cancer Therapies. Adv Exp Med Biol. 2020;1217:123–46.
Ophinni Y, Inoue M, Kotaki T, Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci Rep. 2018;8:7784.
Chen D, Li L, Wang Y, Xu R, Peng S, Zhou L, et al. Ischemia-reperfusion injury of brain induces endothelial-mesenchymal transition and vascular fibrosis via activating let-7i/TGF-βR1 double-negative feedback loop. Faseb j. 2020;34:7178–91.
Liu Z, Li H, Su J, Xu S, Zhu F, Ai J, et al. Numb depletion promotes Drp1-mediated mitochondrial fission and exacerbates mitochondrial fragmentation and dysfunction in acute kidney injury. Antioxid Redox Signal. 2019;30:1797–816.
Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21:93–102.
Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014;31:734–46.
Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2020;27:1709–27.
Funding
This work was supported by grants from the Nature and Science Foundation of China (81730019, 81521003, 82090020) and grants from the Nature and Science Foundation of Guangdong province (2019B1515120075) to Dr. Jing Nie, grant from the Nature and Science Foundation of China (82170700) to Dr. Fengxin Zhu.
Author information
Authors and Affiliations
Contributions
HL and JN designed the experiments and wrote the manuscript. HL, SS, MZ, YC and AX performed in vitro experiments. SS and FZ conducted animal studies, data analysis and manuscript editing. ZH provided reagents and technic supported this study. YM help to checked the spelling and grammar of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Li, H., Shu, S., Zhou, M. et al. NUMB facilitates autophagy initiation through targeting SCFβ-TrCP2 complex. Cell Death Differ 29, 1409–1422 (2022). https://doi.org/10.1038/s41418-022-00930-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00930-3
This article is cited by
-
Ubiquilin-4 induces immune escape in gastric cancer by activating the notch signaling pathway
Cellular Oncology (2023)