This blog post was contributed by Ankit Sethia, PhD, and Timothy Harkins, PhD, at NVIDIA Parabricks, and Olivia Choudhury, PhD, Sujaya Srinivasan, and Aniket Deshpande at AWS.
This blog provides an overview of NVIDIA’s Clara Parabricks along with a guide on how to use Parabricks within the AWS Marketplace. It focuses on germline analysis for whole genome and whole exome applications using GPU accelerated bwa-mem and GATK’s HaplotypeCaller.
Introduction
Next generation sequencing (NGS) platforms have outperformed Moore’s Law when it comes to cost per genome, and short-read sequencing platforms can now readily sequence 96 to 384 whole human genomes in 1-2 days, generating terabytes of data per instrument run [1]. With this increased throughput, whole genome sequencing (WGS) of tens of thousands to hundreds of thousands of human genomes is now common as seen in many national programs to sequence their local populations [2].
Furthermore, WGS and whole exome sequencing (WES) are becoming very important to healthcare systems in addressing undiagnosed issues in the neonatal intensive care units, monitoring cancer treatments, and diagnosing and identifying risk factors for complex disorders like autism or cardiovascular disease. As NGS throughput is increasing, further bringing down the cost per sample, data volumes are increasing exponentially. As a result, data storage, management, and analysis are causing a major bottleneck in the overall workflow and increasing the underlying cost. As state-of-the-art methods allow users to extract more information from their NGS data, making the analytical pipelines more computationally intensive, this bottleneck is getting worse.
To address these computational challenges, the NVIDIA Clara healthcare team has started accelerating and optimizing these genomic analysis pipelines on Graphics Processing Units (GPUs). Traditionally, these chips were used for video-based applications, and as the GPU became more computationally powerful, compute oriented general-purpose workloads started taking advantage of these platforms. GPUs are now the main workhorse of majority of supercomputers and data centers to accelerate key applications.
Clara Parabricks overview
NVIDIA introduced the Clara Parabricks software suite of accelerated genomic analysis to support the three major NGS applications – germline variant analysis, somatic variant detection, and RNA-Seq analysis. The overall goal for the Clara Parabricks software is to provide at least an order of magnitude acceleration in compute time while generating identical outputs and reducing analysis costs. This powerful suite of genomic analysis tools called Clara Parabricks is now available on AWS as an AWS Marketplace AMI. It provides optimal performance for multiple instance types and can be used out of the box for essential bioinformatics needs. Currently, the Clara Parabricks accelerated analysis tools start with a FASTQ file to perform alignment through variant calling and expression analysis, including QC tools for both types of outputs. The suite of 33 tools can be used to support end-to-end workflows for germline, somatic and RNA-Seq pipelines, providing the flexibility to meet the individual needs of most projects.
The figure below shows most of the accelerated tools within the Clara Parabricks software package. Due to the accelerations of the pipelines, users can implement multiple variant callers to extract the most information from their data, and still generate the results in less time and at lower cost than using standard baseline software solutions. For example, GATK’s HaplotypeCaller and Google’s DeepVariant can be used to generate two VCF’s for the same dataset. This enables researchers to either perform a union of both callers to minimize their false negative rates or use the intersection to improve the false positive rates. A standard 30x WGS sample can be processed in less than an hour using both variant callers using p4d.24xlarge instance on AWS. Currently, the Clara Parabricks Pipelines software suite supports Amazon EC2 G4dn, P3 and P4d instances on AWS.
Germline analysis
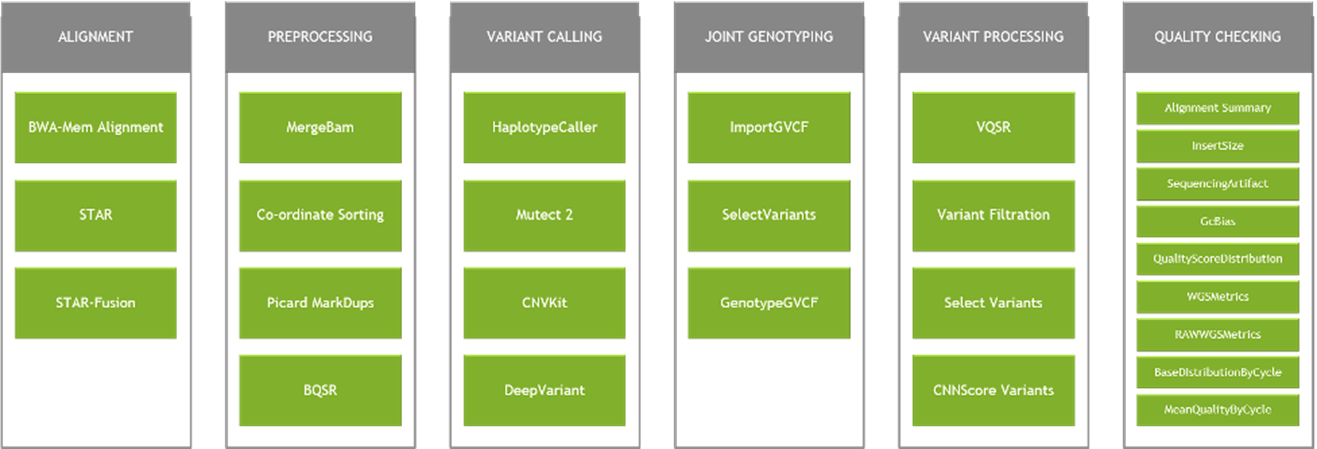
In this blog, we will focus on germline variant detection and associated applications. These are the genetic variants an organism derives from its parents, the inherited variants. This is one of the most popular analyses for DNA sequencing data and is a prerequisite to population scale Genome Wide Association Studies (GWAS). With Clara Parabricks software, a user can go from a 30x human WGS FASTQ to generating a VCF using om using comparable GATK best practices germline analysis (shown below) in as quick as 25 minutes (actual time depends on instance type chosen). The same analysis on one CPU instance with out-of-the-box software can take close to 30 hours. While Clara Parabricks germline analysis also supports Google’s DeepVariant, in this blog, we will focus on the GATK4 best practices pipeline. Similar runtimes can be expected for Google’s DeepVariant as well.
Figure 2: GATK best practices pipeline for germline variant detection.
Running Parabricks Germline Pipeline on AWS
Prerequisites
The prerequisites for running Parabricks on AWS are:
- An AWS account with permission to provision Amazon EC2, Amazon S3, and access AWS Marketplace.
- A VPC with at least a public subnet and a private subnet routed to a NAT Gateway.
Getting Started with the AMI
Step 1: Subscribe to NVIDIA Clara Parabricks Pipelines AMI in AWS Marketplace
NVIDIA Clara Parabricks Pipelines are available as an Amazon Machine Image (AMI) in AWS Marketplace. An AMI provides the necessary information to launch an Amazon EC2 instance. It can also be used to launch multiple instances of the same configuration. To subscribe to this AMI:
- Log in to your AWS account.
- Go to AWS Marketplace and search for “NVIDIA Clara Parabricks Pipelines”.
- Click on Continue to Subscribe.
- Read and accept the terms and conditions.

Once the subscription is complete, the AMI will appear on your list of AWS Marketplace Subscriptions, as shown in Figure 3. To launch an EC2 instance using this AMI:
- Click on Launch new instance.

- Use the pre-set values for Delivery method and Software version and select the Region in which you want to launch the instance.
- Choose an EC2 instance type. The recommended instance type is g4dn.12xlarge, which is selected by default.
- Configure instance detail. For the purpose of this demonstration, you can select the default setting and click on Review and Launch. This will launch a g4dn.12xlarge instance in a default VPC and subnet.
Once the instance is launched and ready to be used, use SSH to log in it.
ssh -i <access-key.pem> user-id@<public-DNS>
Step 3: Download data
cd /mnt/disks/localaws s3 cp s3://genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/30x/HG001.novaseq.pcr-free.30x.R1.fastq.gz .aws s3 cp s3://genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/30x/HG001.novaseq.pcr-free.30x.R2.fastq.gz .aws s3 cp s3://parabricks.sample/parabricks_sample.tar.gz .tar -xvzf parabricks_sample.tar.gz
Step 4: Run Parabricks
To run Parabricks germline pipeline on the above-mentioned dataset, you can use the following command:
cd /mnt/disks/localpbrun germline –ref parabricks_sample/Ref/Homo_sapiens_assembly38.fasta \ –in-fq HG001.novaseq.pcr-free.30x.R1.fastq.gz HG001.novaseq.pcr-free.30x.R2.fastq.gz \ –out-bam 30x.bam \ –out-variants 30x.vcf \ –knownSites parabricks_sample/Ref/Homo_sapiens_assembly38.known_indels.vcf.gz \ –out-recal-file 30x_recal.txt
Benchmark analysis
We use the 30x HG001 hosted on Amazon S3 here for thorough analysis across instance types: s3://genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/30x/
We use 50x, 75x, and 100x WES data for HG001 found here: s3://genomics-benchmark-datasets/google-brain/fastq/novaseq/wes_agilent/
We also use 50x WGS data for HG001 found here: s3://genomics-benchmark-datasets/google-brain/fastq/novaseq/wgs_pcr_free/50x/
We run the Parabricks software across the following instance types: g4dn.12xlarge, g4dn.metal, p3dn.24xlarge and p4dn.24xlarge.
Read the full blog to see benchmarking results and success stories from customers already using Clara Parabricks.
Reminder: You can learn a lot from AWS HPC engineers by subscribing to the HPC Tech Short YouTube channel, and following the AWS HPC Blog channel.