Abstract
The Japanese macaque (Macaca fuscata) is native to the main islands of Japan, except Hokkaido, and is the most northerly living non-human primate. In the Chubu Sangaku National Park of the Japanese Alps, macaques live in one of the coldest areas of the world, with snow cover limiting the availability of preferred food sources. Winter is typically a bottleneck for food availability potentially resulting in marked energy deficits, and mortality may result from famine. However, streams with groundwater upwelling flow during the winter with a constant water temperature of about 5 °C are easily accessible for Japanese macaques to search for riverine biota. We used metabarcoding (Cytochrome c oxidase I) of fecal samples from Japanese macaques to determine their wintertime diet. Here we provide the first robust evidence that Japanese macaques feed on freshwater biota, including brown trout, riverine insects and molluscs, in Chubu Sangaku National Park. These additional food sources likely aid their winter survival.
Similar content being viewed by others
Introduction
Japanese macaques (Macaca fuscata) typically feed on fruits, seeds, mature and young leaves, and flowers, but are also known to consume fungi and terrestrial invertebrates1,2. The fruits available to macaques have been shown to be affected by habitats changing close to streams3. A diverse number of terrestrial invertebrate taxa are also eaten by Japanese macaques1,4. Despite their small size invertebrates provide larger amounts of energy, protein and fat per unit mass than other foods, such as fruit and vegetation5. In winter their preferred foods are not widely available in snowy areas, so Japanese macaques typically consume the bark and buds of woody plants6,7,8, particularly in areas of Japan, like Kamikochi and Shiga Heights9,10,11,12. The diet of Japanese macaques in heavy snow conditions is typically influenced more by resource availability than by diet preference12. Although, invertebrates may represent an alternative food source in winter, typically feeding on terrestrial insects by Japanese macaques is strongly correlated with higher air temperature as the availability of insects increases from June to August13,14. With snow cover inhibiting their preferred food sources during the winter, groundwater influenced streams with no ice cover are easily accessible for Japanese macaques to turn over cobbles and potentially feed on riverine biota. Macaques in Kamikochi have been circumstantially documented to potentially eat aquatic insects during the winter season through binocular observations15. However, from these observations it was not known which species were consumed as some of the taxa documented in the stream are not found in Kamikochi. In this study we used metabarcoding of the Cytochrome c oxidase I (COI) gene to examine fecal samples from Japanese macaques in the mountainous area of Kamikochi in Chubu Sangaku National Park to determine their wintertime diet conclusively. The samples were collected over three separate winter sampling periods.
Results
A total of 1,896 Amplicon Sequence Variants (ASVs) were found from the 38 samples of Japanese macaque feces collected in the winter seasons 2017 to 2019 (see Materials and Methods). To explore the component of the diet sourced from the streams, we then focused only on sequences from freshwater organisms. DNA from fish were present in seven fecal samples, molluscs in nine samples and arthropods with an aquatic life stage in 18 samples (Table 1).
When looking at the entire eukaryotic community, the dominant phylum in the M. fuscata’s feces in 2017 and 2019 was Arthropoda (16–100% relative abundance in each sample) while Chordata was dominant in 2018 (0–100%). Molluscs were also abundant in the feces in 2018 and 2019, but only in nine samples and at a lower relative abundance (2–61%).
The freshwater fish Salmo trutta (brown trout) was definitively identified in 2018 and 2019 from the fecal samples (Table 2). The sequences had 100% sequence similarity to other Salmo trutta entries in the genetic sequence database GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The freshwater mollusc species found in the diet (Potamopyrgus antipodarum, the New Zealand mud snail, and Semisulcospira dolorosa) were identified with > 99% sequence similarity in 2018 and 2019 but were not present in 2017. Freshwater arthropod insect sequences were found in all years with five genera detected. Two genera were Plecoptera, Nemoura fluva and Sweltsa sp., the other three were Conchapelopia sp. (Chironomidae), Tipula sp. (Tipulidae) and Dixa sp. (Dixidae). Nemoura fluva showed over 98% sequence similarity and the other four genera over 90% similarity to sequences in GenBank. One species of freshwater copepod was also found, Mesocyclops leuckrati, with over 99% sequence similarity.
Discussion
To the best of our knowledge, there are no previous published accounts of Japanese macaques definitively eating freshwater animals in streams, including brown trout. Feeding strategies of Japanese macaques change according to seasonal fluctuations in food resources16,17 and typically winter is a bottleneck for food availability18 thereby potentially resulting in a significant energy deficit19. Japanese macaques are at their upper elevation limit (1500 m) in Kamikochi and mortality may result from famine or hypothermia which tends to occur towards the end of winter in March and April20.
Consumption of vertebrates is rare21. Previously, Japanese macaques have been shown to opportunistically capture marine fish, either when being dried22 or washed up on beaches23. Closely related species have been shown to feed on freshwater fish. For example, long tailed macaques actively capture fish from freshwater pools24, and wild chacma baboons from drying desert pools25. Presumably, the Japanese macaques follow a similar approach capturing brown trout in shallow pools along the stream margin although they may opportunistically eat dead fish although none were observed despite extensive wintertime sampling of the benthos. The capture of live fish has been suggested as probably rare in Japanese macaques25 but brown trout were found in seven fecal samples from 2018 and 2019. Unfortunately, using metabarcoding, it is not possible to determine, whether the DNA signal came from different individual fish in each subsequent year.
Animals attached to rocks require less energy expenditure to capture than fish. Although terrestrial insects are a major source of food for Japanese macaques, especially in summer, this is the first confirmed record of aquatic insect larvae and nymphs in their diet. Of the five genera that were found, three (N. fluva and Sweltsa sp., the two stoneflies, and the dipteran Tipula sp.) are relatively large (>1 cm) at the final instar stage. At the time of year the fecal samples were collected, these insects would not be present as adults, in their terrestrial life-cycle stage, so we conclude that they were captured from the freshwater environment. Japanese macaques collecting insects by turning stones over in the stream is a form of extractive foraging26.
Japanese macaques have previously been reported to eat marine mussels27 and terrestrial snails and slugs11,28. The Burmese long-tailed macaque had been shown to use stone tools to crack open shellfish and this foraging task is time intensive29,30. One of the snails Potamopyrgus antipodarum, the New Zealand mud snail eaten at Kamikochi, is an invasive species first identified in Japan in 199031,32 with an average shell size of 0.5 cm. These molluscs are easily removed from the stones and we assume they are swallowed whole and feeding on them does not involve forceful extraction from the shell, as with marine mussels.
Due to their relatively small size, we speculate that the freshwater copepod, Mesocyclops leuckrati was consumed through the Japanese macaques drinking stream water.
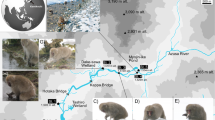
In general, Japanese macaques have a wider home range in winter when food resources are scarce. However, the Kamikochi area lies in a deep valley where the surrounding mountain ranges are >2500 m and have steep inclines, so their home range cannot be expanded. As a result, the population density is exceptionally high, increasing from 90 individuals in two troops in 1990 to 205 individuals in four troops in 20186,33. These Kamikochi macaques are thus forced to overwinter within an extremely harsh environment with high snowfall (mean max depth for 2017–2019 was 930 mm) and low winter temperature (minimums to –20 °C see Fig. 1). These larger populations create additional stress for overwintering survival. An abundance of groundwater upwellings and hot spring inputs from active volcanoes in the Kamikochi region ensures many streams are flowing over the winter without ice cover allowing easy access to the monkeys. Many streams have a stable water temperature year-round (5 to 6 °C) contributing to high biomass of fish and freshwater benthos34 which would be accessible to the Japanese macaques. Hence, the Kamikochi area may be the only environment in Japan where the topographical, geological, and meteorological conditions facilitate this unique feeding of freshwater biota by Japanese macaques to supplement their winter diet.
Methods
Samples were collected by locating fresh feces close to the Azusa River in Kamikochi (Chubu Sangaku National Park 36°15′09.80"N 137°40′02.33"E). Small amounts (ca. 1 g) were sampled from the centre of the feces with a sterilized spatula and placed in a 15 ml tube containing LifeGuard Soil Preservation Solution (QIAGEN, CA, USA). Five samples were collected December 2017, 15 samples in February 2018, and 18 samples in late March/early April 2019. These samples were at least 50 m apart to minimise the chance that fecal samples were from the same individual and in 2018 and 2019 samples were collected at least 7 days apart.
Each step of the molecular analysis (DNA extraction, PCR setup, template addition, electrophoresis and PCR clean-up) was conducted in a separate sterile laboratory dedicated to that step with sequential workflow to ensure no contamination. Each room was equipped with ultra-violet sterilization which was switched on for a minimum of 15 min before and after each use. The PCR set-up and template addition were undertaken in laminar flow cabinets with HEPA filtration.
Each sample was homogenized using sterile beads (2 min, 1,500 RPM; 1600 MiniG Automated Tissue Homogenizer and Cell Lyser, SPEX SamplePrep, NJ, USA) and a subsample (ca. 0.2 g) weighted directly into the first tube of a DNeasy PowerSoil DNA Isolation Kit (QIAGEN, CA, USA). DNA was extracted following manufacturer’s instructions using a robotic workstation (QIAcube, QIAGEN). Samples were frozen (−20 °C) until further processing and negative extraction controls were included in each run to monitor contamination.
As Japanese macaque DNA was potentially more prevalent than dietary organisms during high-throughput sequencing (HTS), a blocking primer approach was used to inhibit DNA amplification of the Japanese macaque during the PCR step35. A blocking primer was designed to overlap with the 3′-end of the forward PCR primer and modified with a C3 spacer. The blocking primer was included at 10 times the concentration of PCR primers during amplification.
For metabarcoding, a segment of approximately 300 bp of the mitochondrial cytochrome oxidase 1 (COI) gene was amplified by PCR using the primers mlCOIintF: 5’-GGW ACW GGW TGA ACW GTW TAY CCY CC-3’ and jgHCO2198: 5’-TAI ACY TCI GGR TGI CCR AAR AAY CA-3′36 and the blocking primer M. fuscata-blocker 5’-ATA TTC CCCCCT AGC AGG AAA CTT CTC C /3SpC3/-3’ designed in this study. PCR amplification was undertaken in a total volume of 50 μL using 25 μL of MyFi™ DNA Polymerase (Bioline, USA), 1 μL of each primer (10 µM), 1 μL of blocking primer (100 µM), 15 µL of DNA-free water, 2 µL of BSA (0.2 mg mL−1; Sigma, USA) and 5 µL of template DNA (ca. 5 ng µL−1). Thermocycling conditions were: 95 °C for 5 min, followed by 40 cycles of 95 °C for 20 s, 52 °C for 20 s, 72 °C for 60 s, and a final extension of 72 °C for 10 min. Two samples of RNA/DNA-free water were used as negative controls following the protocol described above.
PCR products were visualized on 1.5% agarose gel with Red Safe™ DNA Loading Dye (Herogen Biotech, USA) and UV illumination. PCR negatives were run to assess for contamination during the PCR steps. The PCR products were purified, cleaned of primer dimers and normalized using SequalPrep Normalisation plate (ThermoFisher, MA, USA), and submitted to Auckland Genomics (University of Auckland, New Zealand) for library preparation. Sequencing adapters and sample-specific indices were added to each amplicon via a second round of PCR using the Nextera™ Index kit (Illumina Inc., USA). Quality control was undertaken using a bioanalyzer before the library was diluted to 4 nM and denatured. A 15% PhiX spike was used and the final loading concentration was 7 rM. Sequence data were automatically demultiplexed using MiSeq® Reporter (version 2, Illumina Inc.), and forward and reverse reads assigned to samples. Raw sequence reads were deposited in the National Center for Biotechnology Information (NCBI) short read archive under the accession number PRJNA706701.
Raw reads were processed, subsequent to primers being removed with cutadapt37 using the DADA2 package38 within R. Reads were truncated to 221 and 223 bp and filtered with a maxEE (maximum number of “expected errors”) of 2 and 2 for forward and reverse reads respectively (reads not reaching this threshold were discarded). DADA2 constructs a parametric error matrix (based on the first 108 bps in the dataset), the samples are dereplicated and sequence variants for the forward and reverse reads are inferred based on the derived error profiles from the samples. Singletons observed in the inference step are discarded. Subsequently, paired-end reads were merged with a maximum mismatch of 1 bp and a required minimum overlap of 10 bp. Forward and reverse reads, which did not merge were not included in further analysis. Chimeras were removed using the function removeBimeraDenovo. The resulting chimera-checked, merged Amplicon Sequence Variants (ASVs) were used for taxonomic classification using a redundant BOLD database39 supplemented with COI sequences from NCBI within the DADA2 package, which is based on the rdp classifier40 with a bootstrap of 50. The results were parsed into a table using the phyloseq package41, and reads assigned as primates were removed. The differentiation of the sequences from the annelids present in the Japanese macaque’s intestinal tract already from those in the diet was not feasible and thus removed. Negative controls were assessed and the sum of reads from contaminating ASVs was subtracted from the samples.
Phylogenetically annotated COI sequences were used to characterize eukaryotic composition for each fecal sample using the package ggplot242 in R. Sequences of interest (freshwater arthropods, molluscs and fish) were selected and blasted using the BOLD and the NCBI databases for a more accurate taxonomy identification.
Data availability
Raw sequence reads were deposited in the National Center for Biotechnology Information (NCBI) short read archive under the accession number PRJNA706701.
References
Agetsuma, N. Foraging strategies of Yakushima Macaques (Macaca-fuscata Yakui). Int. J. Primatol. 16, 595–609. https://doi.org/10.1007/bf02735283 (1995).
Hill, D. A. Seasonal variation in the feeding behavior and diet of Japanese macaques (Macaca fuscata yakui) in lowland forest of Yakushima. Am. J. Primatol. 43, 305–322 (1997).
Otani, Y. et al. Factors influencing riverine utilization patterns in two sympatric macaques. Sci. Rep. https://doi.org/10.1038/s41598-020-79259-1 (2020).
Maruhashi, T. Feeding behavior and diet of the Japanese monkey Macaca-fuscata-yakui on Yakushima Island, Japan. Primates 21, 141–160. https://doi.org/10.1007/bf02374030 (1980).
Rothman, J. M., Raubenheimer, D., Bryer, M. A. H., Takahashi, M. & Gilbert, C. C. Nutritional contributions of insects to primate diets: Implications for primate evolution. J. Hum. Evol. 71, 59–69. https://doi.org/10.1016/j.jhevol.2014.02.016 (2014).
Hanya, G. et al. Not only annual food abundance but also fallback food quality determines the Japanese macaque density: evidence from seasonal variations in home range size. Primates 47, 275–278. https://doi.org/10.1007/s10329-005-1076-2 (2006).
Nakagawa, N. Determinants of the dramatic seasonal changes in the intake of energy and protein by Japanese monkeys in a cool temperate forest. Am. J. Primatol. 41, 267–288. https://doi.org/10.1002/(sici)1098-2345(1997)41:4%3c267::aid-ajp1%3e3.0.co;2-v (1997).
Tsuji, Y., Ito, T. Y., Wada, K. & Watanabe, K. Spatial patterns in the diet of the Japanese macaque Macaca fuscata and their environmental determinants. Mammal Rev. 45, 227–238. https://doi.org/10.1111/mam.12045 (2015).
Wada, K. & Tokida, E. Habitat utlization by wintering Japanese monkeys Macaca fuscata-fuscata in Shiga Heights Japan. Primates 22, 330–348. https://doi.org/10.1007/bf02381574 (1981).
Suzuki, A. An ecological study of wild Japanese monkeys in snowy area focused on their food habits. Primates 6, 31–71 (1965).
Izawa, K. & Nishida, T. Monkeys living in the northern limits of their distribution. Primates 4, 67–88 (1963).
Enari, H. & Sakamaki-Enari, H. Influence of heavy snow on the feeding behavior of Japanese Macaques (Macaca Fuscata) in Northern Japan. Am. J. Primatol. 75, 534–544. https://doi.org/10.1002/ajp.22128 (2013).
Agetsuma, N. Dietary selection by Yakushima macaques (Macaca-fustcata Yakui): the influence of food availability and temperature. Int. J. Primatol. 16, 611–627. https://doi.org/10.1007/bf02735284 (1995).
Agetsuma, N. & Nakagawa, N. Effects of habitat differences on feeding behaviors of Japanese monkeys: comparison between Yakushima and Kinkazan. Primates 39, 275–289. https://doi.org/10.1007/bf02573077 (1998).
Izumiyama, S. In: High Altitude Primates, Developments in Primatology: Progress and Prospects Vol. 44 (ed N.B. Grow et al.) 153–181 (Springer, New York, 2014).
Go, M. Seasonal changes in food resource distribution and feeding sites selected by Japanese macaques on Koshima Islet, Japan. Primates 51, 149–158. https://doi.org/10.1007/s10329-009-0179-5 (2010).
Hanya, G. Diet of a Japanese macaque troop in the coniferous forest of Yakushima. Int. J. Primatol. 25, 55–71. https://doi.org/10.1023/b:ijop.0000014645.78610.32 (2004).
Sakamaki, H., Enari, H., Aoi, T. & Kunisaki, T. Winter food abundance for Japanese monkeys in differently aged Japanese cedar plantations in snowy regions. Mammal Study 36, 1–10. https://doi.org/10.3106/041.036.0101 (2011).
Enari, H. In: High Altitude Primates. Developments in Primatology, Progress and Prospects (eds N Grow, S Gursky-Doyen, & Krzton A) (Springer, New York, 2014).
Tsuji, Y. & Nakagawa, N. Monkeys of Japan: A Mammalogical Studies of Japanese Macaques (University of Tokyo Press, 2017).
Suzuki, S., Hill, D. A., Maruhashi, T. & Tsukuhara, T. Frog and Lizard-eating behaviour of wild Japanese Macaques in Yakushima, Japan. Primates 31, 421–426 (1990).
Watanabe, K. Fish: a new addition to the diet of Japanese macaques on Koshima Island. Folia Primatol. 52, 124–131. https://doi.org/10.1159/000156391 (1989).
Leca, J. B., Gunst, N., Watanabe, K. & Huffman, M. A. A new case of fish-eating in Japanese macaques: Implications for social constraints on the diffusion of feeding innovation. Am. J. Primatol. 69, 821–828. https://doi.org/10.1002/ajp.20401 (2007).
Stewart, A. M. E., Gordon, C. H., Wich, S. A., Schroor, P. & Meijaard, E. Fishing in Macaca fascicularis: a rarely observed innovative behavior. Int. J. Primatol. 29, 543–548. https://doi.org/10.1007/s10764-007-9176-y (2008).
Hamilton, W. J. & Tilson, R. L. Fishing Baboons at desert waterholes. Am. J. Primatol. 8, 255–257. https://doi.org/10.1002/ajp.1350080308 (1985).
Tamura, M. Extractive foraging on hard-shelled walnuts and variation of feeding techniques in wild Japanese macaques (Macada fuscata). Am. J. Primatol. 82, e23130 (2020).
Iwamoto, T. A bioeconomic study on a provisioned troop at a Japanese monkeys Macada fuscata-fuscata at Koshima Islet Miyazaki. Primates 15, 241–262. https://doi.org/10.1007/bf01742286 (1974).
Tsuji, Y. & Takatsuki, S. Effects of a typhoon on foraging behavior and foraging success of Macaca fuscata on Kinkazan Island, Northern Japan. Int. J. Primatol. 29, 1203–1217. https://doi.org/10.1007/s10764-008-9293-2 (2008).
Gumert, M. D. & Malaivijitnond, S. Marine prey processed with stone tools by burmese long-tailed macaques (Macaca fascicularis aurea) in intertidal habitats. Am. J. Phys. Anthropol. 149, 447–457. https://doi.org/10.1002/ajpa.22143 (2012).
Tan, A., Tan, S. H., Vyas, D., Malaivijitnond, S. & Gumert, M. D. There is more than one way to crack an oyster: identifying variation in burmese long-tailed Macaque (Macaca fascicularis aurea) stone-tool use. PLoS ONE https://doi.org/10.1371/journal.pone.0124733 (2015).
Urabe, M. The present distribution and issues regarding the control of the exotic snail Potamopyrgus antipodarum in Japan. Jpn. J. Limnol. 68, 491–496 (2007).
Hamada, K. T. Y. & Urabe, M. Survey of mitochondrial DNA haplotypes of Potamopyrgus antipodarum (Caenogastropoda: Hydrobiidae) introduced into Japan. Limnology 14, 223–228 (2013).
Izumiyama, S., Mochizuki, T. & Shiraishi, T. Troop size, home range area and seasonal range use of the Japanese macaque in the Northern Japan Alps. Ecol. Res. 18, 465–474. https://doi.org/10.1046/j.1440-1703.2003.00570.x (2003).
Milner, A. M., Docherty, C., Windsor, F. M. & Tojo, K. Macroinvertebrate communities in streams with contrasting water sources in the Japanese Alps. Ecol. Evol. 10, 7812–7825. https://doi.org/10.1002/ece3.6507 (2020).
Vestheim, H. & Jarman, S. N. Blocking primers to enhance PCR amplification of rare sequences in mixed samples: a case study on prey DNA in Antarctic krill stomachs. Front. Zool. https://doi.org/10.1186/1742-9994-5-12 (2008).
Leray, M. et al. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. https://doi.org/10.1186/1742-9994-10-34 (2013).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3. doi:https://doi.org/10.14806/ej.17.1.200 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581. https://doi.org/10.1038/nmeth.3869 (2016).
Ratnasingham, S. & Hebert, P. D. N. BOLD: the barcode of life data system (www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi:https://doi.org/10.1111/j.1471-8286.2007.01678.x (2007).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. https://doi.org/10.1128/aem.00062-07 (2007).
McMurdie, P. J. & Holmes, S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE https://doi.org/10.1371/journal.pone.0061217 (2013).
Wickham, H. ggplot2: elegant graphics for data analysis 2nd edn. (Springer, 2016).
Acknowledgements
We thank Prof. Keisuke Suzuki of Shinshu University and the staff of the Mountain Environmental Science Center of Shinshu for the meteorological observations. Thanks to Dr. Tomoya Suzuki, Dr. Koki Yano and Dr. Miquel Vall‐llosera Camps for assistance with collecting field samples. We are grateful to Prof. Shigeyuki Izumiyama, Dr. Takuya Matsumoto, and Mr. Masaya Tamura for their valuable knowledge of Japanese macaques.
Funding
Japan Society for Promotion of Science Long Term Fellowship L16553 (AM). Japan Society for Promotion of Science 17H02033, 20H031390 (KT). Shinshu University Postdoctoral Fellowship (CD). Institute of Mountain Science Shinshu University (KT). EU project HiFREQ H2020-MSCA-RISE-2016-734317 (AM, CD).
Author information
Authors and Affiliations
Contributions
Conceptualization: AM, KT, CD; Investigation: AM, KT, CD, SW, LB, MT; Funding acquisition: AM, KT, CD; Project administration: AM, KT; Writing: review and editing: AM, KT, CD, SW, LB, MT.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Milner, A.M., Wood, S.A., Docherty, C. et al. Winter diet of Japanese macaques from Chubu Sangaku National Park, Japan incorporates freshwater biota. Sci Rep 11, 23091 (2021). https://doi.org/10.1038/s41598-021-01972-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01972-2
This article is cited by
-
Behavior of snow monkeys hunting fish to survive winter
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.