Abstract
Human coronaviruses cause a wide spectrum of disease, ranging from mild common colds to acute respiratory distress syndrome and death. Three highly pathogenic human coronaviruses — severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus and SARS-CoV-2 — have illustrated the epidemic and pandemic potential of human coronaviruses, and a better understanding of their disease-causing mechanisms is urgently needed for the rational design of therapeutics. Analyses of patients have revealed marked dysregulation of the immune system in severe cases of human coronavirus infection, and there is ample evidence that aberrant immune responses to human coronaviruses are typified by impaired induction of interferons, exuberant inflammatory responses and delayed adaptive immune responses. In addition, various viral proteins have been shown to impair interferon induction and signalling and to induce inflammasome activation. This suggests that severe disease associated with human coronaviruses is mediated by both dysregulated host immune responses and active viral interference. Here we discuss our current understanding of the mechanisms involved in each of these scenarios.
Similar content being viewed by others
Introduction
Coronaviruses can cause highly pathogenic diseases in humans, as evidenced by three major outbreaks within the past two decades. The severe acute respiratory syndrome (SARS) epidemic first occurred in 2002 and was later eradicated, but caused around 8,000 cases of disease, with mortality of roughly 10%1. The Middle East respiratory syndrome (MERS) epidemic is still ongoing, predominantly in the Middle East, with more than 2,000 cases and 800 deaths occurring since MERS coronavirus (MERS-CoV) was first identified in 2012 (ref.2). The current COVID-19 pandemic, caused by SARS coronavirus 2 (SARS-CoV-2), has resulted in more than 242 million cases, with a death toll of more than 4.9 million people (as of 25 October 2021 (ref.3)). In light of the continuous efforts to understand these highly pathogenic coronaviruses, scientists have described the immunopathological nature of immune responses to these coronaviruses. Highlighting this, host immune dysregulation has been shown to contribute to disease severity and to determine disease outcome in these infections. However, abundant evidence has also demonstrated that active immune evasion by viral proteins encoded by SARS-CoV, MERS-CoV and SARS-CoV-2 further contributes to the dysregulation of host immune processes. In this Perspective, we summarize current knowledge of host immune dysregulation during infection with SARS-CoV-2 and related human coronaviruses. We also describe and discuss the immunopathological host response and the mechanistic role of viral proteins in the active manipulation of the host immune system.
Dysregulated interferon responses
Conflicting data on the role of interferons in coronavirus infection
Interferons are a group of antiviral cytokines that are induced during viral infections. The antiviral functions of interferons can be largely attributed to interferon-stimulated genes (ISGs). ISGs are upregulated by interferons during viral infection to perform critical effector functions for virus containment and clearance. Antiviral effector functions of ISGs include, but are not limited to, inhibiting virus entry, replication, translation and egress4. In addition to direct antiviral functions, ISGs also regulate adaptive immune responses by recruiting and directing the differentiation of immune cells4. The protective roles of the interferon pathway have been evidenced by studies reporting that patients with COVID-19 who have genetic defects in interferon signalling pathways or have autoantibodies to interferon have poor disease outcomes. Ten per cent of patients with life-threatening COVID-19 had serum autoantibodies to type I interferon, while 3.5% of patients with severe COVID-19 had genetic defects at loci involved in Toll-like receptor 3 (TLR3)-dependent and interferon-regulatory factor 7 (IRF7)-dependent expression and amplification of type I interferon5,6. In this regard, the benefits of interferon in combination with other antiviral molecules as therapeutic treatments for coronavirus infection have been tested in clinical trials. However, these studies have shown inconsistent results, probably reflected by the timing of interferon administration and thus highlighting the limitations of the direct use of interferons as therapeutic options7,8,9,10,11. The conventional idea that the interferon response is protective in coronavirus infection has been challenged by some studies that suggest pathological roles of the interferon response, especially at the peak of virus replication. For example, this understanding of the role of the interferon response has been complicated by the different clinical outcomes observed in patients. In a clinical study of 40 patients with SARS, robust early interferon and ISG expression were hallmarks of severe disease12. Most patients in the study resolved the interferon response as part of disease resolution and their production of antibodies to SARS-CoV rose, leading to better outcomes. However, patients with a persistent interferon response showed lower oxygen saturation levels, lower levels of antibodies to SARS-CoV and poor clinical outcomes. This study suggests that persistent interferon responses precluded the switch from innate to adaptive immunity as SARS progressed, resulting in severe disease in some patients12. In a longitudinal study of clinical samples from patients with COVID-19, elevated interferon expression was detected in the blood only of patients with severe disease, and elevated levels of interferons correlated with disease severity and mortality, in support of a pathological role of interferon13. However, in other studies, early interferon responses were observed and were found to later resolve in patients with mild-to-moderate COVID-19, whereas upregulation of interferon was not seen in patients with severe disease14,15. It is not surprising to observe different temporal expression patterns of interferon in relation to disease severity in these different studies, as clinical samples were collected from patients with variable demographic profiles and at different times relative to disease progression, complicating the analysis and interpretation in clinical settings.
Contrasting results have also been obtained in studies of the role of interferon signalling in SARS-CoV-infected mice. Serial passaging of human isolates of SARS-CoV in BALB/c mice resulted in the emergence of six mutations throughout the virus genome that are important for pathogenesis in mice. The resulting mouse-adapted virus caused lethal disease upon infection16. Similar strategies have also been used to generate mouse-adapted virus for MERS-CoV and SARS-CoV-2 (refs17,18). BALB/c mice infected with a lethal dose of a mouse-adapted version of SARS-CoV (SARS-MA15) were protected from fatal disease when interferon-α/β (IFNα/β) receptor (IFNAR) was blocked or genetically depleted16,19. The levels of virus detected in the lungs of SARS-MA15-infected wild-type mice and Ifnar1−/− BALB/c mice were largely comparable at all times during the course of the experiment. The protective effects of genetic knockout of IFNAR in BALB/c mice were attributed to reduced infiltration of inflammatory monocytes and macrophages into the lungs in comparison with wild-type mice, suggesting that the detrimental role of interferon signalling in these mice is immunopathological in nature and independent of virus replication19. By contrast, other studies using 129 and C57BL/6 mice showed that genetic depletion of signal transducer and activator of transcription 1 (STAT1), which drives signalling downstream of IFNAR, resulted in elevated levels of virus and more significant pathological changes in the lungs, along with higher mortality20,21. STAT1-deficient mice failed to control initial replication of SARS-MA15 owing to impaired type I interferon/type III interferon signalling20. In addition, STAT1 was found to be involved in wound repair in an interferon-independent manner, which may represent an additional role for STAT1 in host immunity21. These results in patients and animal models reveal the complex dynamics of host interferon signalling in determining disease outcomes during coronavirus infection and are consistent with the notion that dysregulated interferon responses contribute to severe disease.
Timing of type I interferon/type III interferon response relative to disease onset
On the basis of the preceding discussion, the precise nature (protective or detrimental) of the host interferon response to coronavirus infection is still subject to debate. Also, as pointed out earlier herein, it is likely that the temporal kinetics of interferon expression differs among individuals owing to factors such as host genetics and initial viral dose, thereby contributing to the seemingly disparate effects of interferon on clinical outcomes. Therefore, continuing efforts to understand the dynamics of the host interferon response and how this determines disease protection or aggravation are warranted. Clinical studies have revealed a possible explanation for the variability in disease outcome based on the kinetics of the interferon response. Longitudinal studies of severe cases of SARS and COVID-19 revealed delayed and sustained upregulation of interferon responses for extended periods and without resolution in comparison with mild-to-moderate cases12,13,14. These results corroborate findings in murine models of SARS and MERS. Virus titres in the lungs of BALB/c mice infected with SARS-MA15 peaked before the peak of interferon expression, and the detrimental effects of interferon signalling in BALB/c mice were attributed to this delay in interferon expression relative to the peak of virus titres19. This was supported by the protective role of interferon treatment provided before the peak of virus replication, but not when interferon was provided after the peak of virus replication19. The therapeutic role of interferon in MERS was also tested in a murine model where exons 11–14 of the gene encoding human dipeptidyl peptidase 4 (hDPP4), the receptor for MERS-CoV, were knocked in to C57BL/6 mice (hDPP4-KI mice)18,22. Similarly to the SARS study, early interferon treatment of MERS-CoV-infected mice converted an otherwise uniformly lethal infection to a sublethal infection, while delaying interferon treatment until after the peak of virus replication exacerbated disease and resulted in significantly higher mortality22. In SARS-CoV-2 experimentally infected animal models, treatment with interferon or interferon receptor agonists before infection offered protection against severe disease23,24,25. In addition, hamsters challenged with SARS-CoV-2 were protected by prophylactic interferon treatment (1 day before infection) or early interferon treatment (1 day after infection), while late interferon treatment (3 days after infection) conferred no protection26. Thus, the apparently contradictory results of exogenous interferon treatment in different mouse strains could potentially be explained by different replication kinetics of the virus between these mouse strains. In support of this, endogenous interferon signalling was protective in mice infected with a mouse-adapted version of MERS-CoV (MERS-MA30) but was pathogenic in SARS-MA15-infected mice. SARS-MA15 replicated to peak titres at 16 hours after infection in BALB/c mice, whereas titres of MERS-MA30 peaked at 2 days after infection in hDPP4-KI mice. It was therefore proposed that the timing of interferon production relative to the peak virus titres, which is specific to the host and virus in context, dictates the nature of host endogenous interferon signalling and the outcome of exogenous interferon treatment19,22.
Several studies have attempted to delineate the pathological roles of interferon signalling in mice infected with human coronaviruses. Pathological consequences of endogenous or exogenous type I interferon were shown to be mediated in part by the infiltration of inflammatory monocytes and macrophages into the lungs of infected mice. In hDPP4-KI mice infected with a sublethal or lethal dose of MERS-MA30, mice that received interferon exogenously at early times after infection had significantly fewer activated monocytes and macrophages in their lungs than infected mice that were not treated with interferon. Moreover, the severe disease observed in SARS-MA15-infected BALB/c mice or in mice that received interferon coincident with peak virus titres was associated with notably higher numbers of activated monocytes and macrophages in the lungs19,22. The detrimental effects of enhanced inflammatory monocyte infiltration were nullified by treatment with a monocyte-depleting anti-CCR2 antibody, confirming the key role of these cells in exacerbating disease19,22. In addition, human autopsy studies and analyses of peripheral blood mononuclear cells also identified elevated numbers of monocytes and macrophages in the lungs and blood of patients with severe COVID-19 (refs27,28,29). Transcriptomic analyses revealed a robust interferon gene signature in macrophages from the airways of patients with COVID-19, supporting the idea that monocytes and macrophages mediate the immunopathological effects of interferon. For example, interferon signalling resulted in the upregulation of pro-inflammatory cytokines in macrophages in these studies, possibly contributing to immunopathological changes13,30,31,32,33. Additional studies in mice showed that type I interferon/type III interferon inhibited regeneration of the lung epithelia, in part by inducing the expression of the tumour suppressor protein p53, which impaired the proliferation of alveolar type II cells and differentiation of basal cells during recovery34,35. Consistent with these observations, treatment of mice with poly(I:C), a Toll-like receptor 3 (TLR3) agonist, resulted in inhibition of lung epithelial repair34. Together, these results suggest that elevated expression of type I and type III interferons in the lower airways of patients with severe COVID-19 during the late acute period, after virus titres have peaked, contributes to poor outcomes through interferon-mediated inhibition of lung epithelial regeneration36. These results indicate that a temporally dysregulated type I interferon/type III interferon response can impair all parts of the host response to infection. They also suggest that localized effects of interferon signalling need to be considered as the role of interferon in COVID-19 is further investigated35.
Interferon antagonism by viral proteins encoded by coronaviruses
In addition to the host factors that contribute to a dysregulated interferon response, coronaviruses exhibit variable sensitivity to interferon, and SARS-CoV, MERS-CoV and SARS-CoV-2 all encode interferon antagonists that actively interfere with host interferon induction and/or signalling37,38,39,40. In particular, MERS-CoV and SARS-CoV-2 are more sensitive to interferon than SARS-CoV, and these two viruses inhibit interferon induction to a greater extent than SARS-CoV. However, SARS-CoV and SARS-CoV-2 appear to inhibit IFNAR signalling to a greater extent than MERS-CoV37,40.
Viral proteins encoded by coronaviruses can be broadly classified into three groups: structural proteins, non-structural proteins and accessory proteins (Fig. 1). Each group of proteins is responsible for specific functions of the viral life cycle. For example, non-structural proteins are critical for viral RNA transcription and replication by forming the replication–transcription complex. Structural proteins include the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, which are essential for the formation of virions. Accessory proteins are dispensable for the viral life cycle but are important for immunoevasive activities. With the goal of identifying interferon antagonists encoded by SARS-CoV-2, in vitro screening using singly transfected SARS-CoV-2 viral proteins was performed. The results suggested that non-structural protein 1 (nsp1), nsp3, nsp12, nsp13, nsp14, nsp15, nsp16, open reading frame 3 (ORF3) protein, ORF6 protein, M protein and N protein are interferon antagonists that suppress interferon expression in various stimulation conditions that mimic SARS-CoV-2 infection41,42,43 (Fig. 2).
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) encodes three types of viral proteins. Sixteen non-structural proteins are encoded by open reading frame 1a (ORF1a) and ORF1b, which make up more than two-thirds of the genome. Non-structural protein 1 (nsp1)–nsp11 are encoded by polyprotein 1a (pp1a). nsp12–nsp16 are expressed in pp1ab only when a ribosomal frameshift occurs at the junction of nsp11 and nsp12, which occurs approximately 25% of the time136. Genes encoding the structural proteins (spike (S), envelope (E), membrane (M) and nucleoprotein (N) proteins) and accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8 and ORF9b proteins) are located downstream of ORF1a/1b. The S protein is responsible for viral entry by interacting with the host cell-expressed receptor angiotensin-converting enzyme 2 (ACE2). The S1 subunit of the S protein binds ACE2, while the S2 subunit triggers fusion. The S1 and S2 subunits are separated by a furin cleavage site, which is not found in SARS-CoV. The accessory proteins of SARS-CoV-2 are dispensable for replication but are crucial in mediating immune evasion. CT, cytoplasmic tail; FP, fusion peptide; HR, heptad repeat; L, leader sequence; NTD, amino-terminal domain; RBD, receptor-binding domain; TM, transmembrane domain.
Coronavirus RNA in the cytoplasm is sensed by the cytoplasmic RNA sensors RIG-I and MDA5. Sensing of viral RNA triggers conformational changes in these sensors and results in the recruitment of downstream effector proteins. MAVS interacts with RIG-I or MDA5 through CARD domains to recruit the downstream kinases TBK1 and IKKε for phosphorylation of interferon-regulatory factor 3 (IRF3) and IRF7. MAVS activation also recruits TNF receptor-associated factor 6 (TRAF6), which serves as an adaptor for the IKK complex (NEMO, IKKα and IKKβ). The IKK complex phosphorylates NF-κB canonical inhibitor, IκB, which results in IκB degradation and activation of NF-κB. IRF3, IRF7 and NF-κB translocate to the nucleus and interact with the corresponding positive regulatory domain (PRD). IRF3 and IRF7 bind PRD I/PRD III, NF-κB binds PRD II, and AP-1 (a heterodimer of JUN and ATF2) binds PRD IV on the interferon-β (IFNβ) promoter to form the interferon enhanceosome for induction of IFNβ expression. IFNβ is secreted and interacts with the IFNα/β receptor (IFNAR; comprising the IFNAR1 and IFNAR2 subunits) in an autocrine or a paracrine manner. Binding of interferon to IFNAR activates the signal transducer and activator of transcription 1 (STAT1) and STAT2 kinases, Janus kinase 1 (JAK1) and the tyrosine kinase TYK2. Phosphorylated STAT1 and STAT2 associate with IRF9 to form ISGF3, which translocates to the nucleus and interacts with the interferon-stimulated response element (ISRE) promoter to drive the expression of downstream interferon-stimulated genes (ISGs). ISGs perform different antiviral functions. The example depicted in the figure is the 2′-5′-oligoadenylate synthetase (OAS)–RNase L pathway. OAS interacts with viral RNA and catalyses the formation of 2′-5′-oligoadenylate (2-5A) from ATP. 2-5A is a secondary messenger that activates RNase L to drive viral RNA degradation. Viral proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and related coronaviruses shown in the figure interfere with interferon production and signalling at different steps. Viral proteins (depicted in red) are from SARS-CoV-2 unless otherwise specified. MERS-CoV, Middle East respiratory syndrome coronavirus; N, nucleocapsid protein; nsp, non-structural protein; ORF, open reading frame protein.
Conserved interferon-antagonizing functions among viral proteins encoded by coronaviruses highlight the important role of interferon antagonism in coronavirus evolution. As an example of this, SARS-CoV ORF6 protein was shown to inhibit interferon signalling by binding karyopherin subunit-α2 (ref.44), which impeded STAT1 nuclear translocation. In the case of SARS-CoV-2, ORF6 protein hijacks the nuclear pore complex to block STAT1 nuclear translocation38, while ORF9b protein interacts with mitochondrial import receptor subunit TOM70 to suppress interferon expression45. In another example, nsp16 is a 2′-O-methyltransferase found in all coronaviruses that inhibits the recognition of viral RNA by intracellular helicases46. nsp16 functions by methylating the 2′-hydroxy group of the first base to form a cap 1 structure to mimic cellular mRNA. In addition, the N protein was shown to undergo liquid–liquid phase separation with RNA to impede interferon expression by preventing MAVS aggregation47. One caveat regarding the screening of interferon antagonists is that most studies thus far have used tissue culture cells with ectopic expression of the protein of interest. In many of these assays, viral proteins were expressed singly in the absence of other viral proteins and often at supraphysiological levels. Results obtained from these experiments should be interpreted with caution, and these results need to be verified using recombinant viruses genetically depleted of the protein of interest. For some proteins, such as the M and N proteins, this is not feasible since they are essential for virus growth. In these cases, it may be possible to perform mutational analyses to identify specific amino acids important for immune evasion but not virus viability. Another key issue that should be addressed is that blood type I interferon levels are low in patients with severe COVID-19, even though these anti-interferon effects would be expected primarily to inhibit interferon production by infected cells and not bystander cells.
Increase in pro-inflammatory mediators
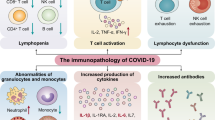
The initial innate inflammatory response to coronaviruses facilitates the rapid recruitment of immune cells to the main site of infection (the lungs) and the subsequent activation, differentiation and proliferation of these cells48. Despite enabling pathogen clearance, this immune response may result in immunopathological changes if unchecked. Multiple reports have indicated pathologically heightened inflammatory responses in patients infected with SARS-CoV, MERS-CoV or SARS-CoV-2 and in animal models of these infections. This was demonstrated by the detection of elevated levels of cytokines and chemokines — including IL-1β, IL-6, IL-8 and tumour necrosis factor (TNF) — in the blood during infection, and the presence of immune cells in the lungs of deceased patients13,15,49,50,51,52. High levels of these inflammatory mediators are also correlated with disease severity13,15,49,50.
Dysregulated inflammatory response alters immune landscape
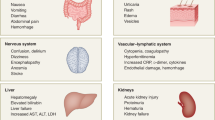
The precise mechanisms of immunopathology in COVID-19 remain elusive. Sustained IL-6 and TNF production in patients infected with SARS-CoV-2 correlates with reduced monocyte maturation. As a result, MHC class II antigen (HLA-DR) expression on circulating monocytes was reduced, resulting in cells less able to present antigen53,54. In addition, these changes in monocyte maturation are accompanied by depletion of natural killer cells, CD4+ T cells and B cells. HLA-DR expression on monocytes and total lymphocyte counts were partially restored with tocilizumab (a monoclonal antibody to IL-6 receptor) treatment53, suggesting a role of sustained IL-6 production in altering the immune landscape. In addition, multisystem inflammatory syndrome in both child and adult patients with a history of COVID-19 was characterized by elevated blood levels of IL-1β, IL-6, IL-8 and IL-10 (ref.55), suggesting a role for the inflammatory response in pathogenesis. Multisystem inflammatory syndrome in children and adults has cardiac, renal, respiratory, haematological, gastrointestinal, dermatological and neurological manifestations, and fever, neutrophilia, lymphopenia and elevated levels of C-reactive protein, fibrinogen, ferritin, IL-6 and D dimer have been detected in laboratory tests55. Animal studies have also indicated a role of elevated levels of inflammatory cytokines in mediating immunopathology in coronavirus infections19,22.
Coronavirus-encoded viral proteins activate inflammasomes and NF-κB signalling
NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasomes are activated in SARS-CoV-2-infected human monocytes and in patients with COVID-19 (refs56,57). NLRP3 activation results in caspase 1 activation and cleavage of IL-1β and IL-18 into their active forms58 as well as the initiation of pyroptosis, a highly inflammatory form of cell death. Several SARS-CoV proteins can activate the NLRP3 inflammasome. The SARS-CoV E protein was shown to possess calcium ion channel activity that activates NLRP3 (ref.59). ORF3a protein of SARS-CoV was demonstrated to cause NLRP3 maturation by enhancing ubiquitination of the NLRP3 adaptor protein ASC by TNF receptor-associated factor 3 (TRAF3)60. ORF8 protein of SARS-CoV also activated NLRP3 through direct interaction61. On the basis of their similarity to SARS-CoV proteins, analogous SARS-CoV-2 proteins are also likely to activate NLRP3 in these ways.
SARS-CoV infection activated NF-κB signalling62, and a hallmark of severe COVID-19 is upregulation of NF-κB-dependent inflammatory molecules, such as IL-1, IL-6, IL-8 and TNF15. In addition to its role in inflammasome activation, the E protein of SARS-CoV induced NF-κB activation62. Despite clear evidence that inflammasome-dependent and NF-κB-dependent cytokines and chemokines induced by SARS-CoV-2 are critical for COVID-19 immunopathogenesis, not much is known about the viral components responsible for such activation. ORF3a, ORF7a, M and N proteins have been reported to induce inflammatory cytokine expression by activating NF-κB in tissue cultures63,64. However, further analysis is warranted to verify how SARS-CoV-2 induces these inflammatory changes with authentic virus through reverse genetics.
Complement-mediated immunopathology in coronavirus infections
The complement system is critical for efficient recognition and elimination of pathogens. However, similarly to the interferon and inflammatory responses, unchecked activation of the complement system can contribute to severe disease. Complement is considered a major component in the endotheliitis and thrombosis observed in COVID-19 (ref.65). Complement activation by a coronavirus was first evident in mice infected with SARS-CoV. Deposition of complement activation products was observed in the lungs of C57BL/6J mice as early as 1 day after infection. C57BL/6J mice genetically deficient in C3 complement protein expression (C3−/− mice) showed significantly improved clinical outcomes after SARS-MA15 infection. SARS-MA15-infected C3−/− mice developed milder inflammatory responses as indicated by fewer neutrophils and inflammatory monocytes in the lungs and diminished levels of inflammatory cytokines and chemokines in the lungs and sera compared with C57BL/6J control mice66. Immunopathogenic complement activation was also observed in mice infected with MERS-CoV, with ameliorated clinical disease when C5a receptor activation was pharmacologically blocked67. Evidence of complement activation in patients with COVID-19, including deposition of the components (C5b, C6, C7, C8 and C9) of the membrane attack complex, C4d and C5a in the microvasculature, lungs, skin and kidney, has been documented68,69,70,71. In particular, the membrane attack complex was associated with respiratory failure70. All these results suggest a role of the complement system in enhancing disease as a result of coronavirus infection.
Despite data showing the involvement of the complement system in enhancing disease, how the complement system is activated and how the complement system mediates pathogenesis during coronavirus infection needs further investigation. Some evidence suggests that neutrophils play a role in complement-mediated injury. It has been proposed that SARS-CoV-2-induced inflammatory responses result in aberrant neutrophil activation and neutrophil extracellular trap formation52,68,72. The anaphylatoxins C3a and C5a, which are complement activation products, promote an inflammatory state by activating neutrophils and other immune cells68,73. Activated neutrophils and neutrophil extracellular traps then interact with the complement system, amplifying the complement activation cascade68. In addition, it is postulated that neutrophil extracellular traps activate coagulation and prothrombotic pathways via the contact system, contributing to arterial and venous thrombosis and coagulopathy, which are observed in many patients with severe disease68,74. A more comprehensive understanding of the role of complement activation in severe COVID-19 is likely to result in additional therapeutic modalities.
Adaptive immunity to coronaviruses
Adaptive immunity is critical for virus clearance and is dysregulated in patients with severe COVID-19. Coronavirus-specific CD4+ T cells, CD8+ T cells, B cells and antibodies have been identified in patients during acute disease and convalescence75,76,77,78,79,80 and were shown to be protective and required for virus clearance in murine models of SARS, MERS and COVID-19 (refs81,82). In mouse models of SARS-CoV, immunization of naive mice with dendritic cells (DCs) pulsed with immunodominant SARS-CoV peptides protected mice from subsequent lethal challenge83. In addition, depletion of CD4+ T cells resulted in reduced immune cell recruitment to the lungs and lower levels of neutralizing antibody production, which coincided with enhanced disease78. Similar results were seen in mice infected with MERS-CoV or SARS-CoV-2 (refs81,83,84,85). In patients with COVID-19, virus-specific T cells respond to stimulation with several peptides across the viral genome, while SARS-CoV-2-specific antibodies target multiple viral proteins75,77,80, denoting the diverse repertoire of viral targets recognized by these cells. In addition, lymphopenia is correlated with severe disease and could contribute to ineffective virus clearance13,15,49,50. We describe in the following subsections the current knowledge of adaptive immunity in patients and animals experimentally infected with SARS-CoV-2 and related coronaviruses and discuss potential evasion mechanisms used by the viruses.
Cellular immune responses and dysregulation in coronavirus infection
Highly pathogenic respiratory coronaviruses, such as SARS-CoV, SARS-CoV-2 and MERS-CoV, infect the lower respiratory tract, but of these three pathogenic human coronaviruses, only SARS-CoV-2 infects the upper airways and nasopharyngeal cavity86,87,88. After initial infection, lung-resident DCs acquire exogenous viral antigens from infected epithelial cells and migrate to the lung-draining lymph nodes, where antigens are presented to naive T cells89. Activated T cells then migrate to the lungs, where they aid in virus clearance (Fig. 3). Disrupting any steps of this process could lead to poor T cell activation and responses. Studies of mice infected with SARS-CoV demonstrated an impact of the lung environment in these processes. In one study, DC migration from the lungs to the draining lymph nodes was impaired by inhibitory alveolar macrophages, leading to suboptimal T cell activation. T cell activation by DCs was restored when alveolar macrophages were depleted in vivo or activated in vitro with a TLR agonist before transfer back to infected mice90. Another example was shown in aged mice, where lung DC migration and subsequent T cell activation were diminished owing to elevated levels of an eicosanoid, prostaglandin D2 (PGD2), and its upstream phospholipase, PLA2G2D, relative to young mice91,92. In aged mice, PGD2 acted on its receptor, PGD2 receptor 1 (DP1), on CD11c+ DCs and suppressed DC migration to the draining lymph nodes, resulting in diminished T cell activation and poor disease outcome relative to young mice. Specifically blocking DP1 signalling on CD11c+ DCs resulted in enhanced DC migration from the lungs to the draining lymph nodes and improved antiviral T cell responses, which partially protected mice from lethal infection with SARS-CoV91. Similar results have also been shown for SARS-CoV-2 (ref.93). These results align well with clinical data indicating that advanced age is a risk factor for SARS, MERS and COVID-19 (refs94,95).
Airway epithelial cells are infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or related coronaviruses. Respiratory dendritic cells (rDCs) acquire viral antigens from infected epithelial cells and process antigens into peptides for MHC loading. Activated rDCs with MHC–peptide complexes present on the cell surface migrate to the lung-draining lymph nodes and present these MHC–peptide complexes to naive T cells. Together with other necessary co-stimulatory signals, T cells are activated upon T cell receptor engagement with the MHC–peptide complexes presented by rDCs. Activated T cells undergo rapid proliferation and migrate to the site of infection for virus clearance. Initial migration of rDCs from the site of infection to the draining lymph nodes (DLNs) is impaired by elevated prostaglandin D2 (PGD2)–PGD2 receptor 1 (DP1) signalling mediated by increased expression of a phospholipase (PLA2G2D) in rDCs in an age-dependent manner91,92. Impaired rDC migration to the draining lymph nodes results in suboptimal T cell activation and poor clinical outcomes in experimentally infected animals.
In addition to studies of experimentally infected animals, evidence indicating the protective role of cellular immune responses in COVID-19 has been described in reports characterizing and comparing T cell responses in asymptomatic, mildly symptomatic and severely symptomatic individuals during acute infection and convalescence. Le Bert et al. compared T cell responses in asymptomatic and symptomatic patients with COVID-19 and identified more robust expression of the T cell effector molecules IL-2 and IFNγ in asymptomatic patients96. Longitudinal studies revealed that early induction, emergence of functional SARS-CoV-2-specific T cells targeting diverse epitopes and prolonged contraction of T cell responses are characteristics observed in patients with mild diseases97,98,99. Tissue-resident T cells in the airways of patients with COVID-19 exhibit functionally protective phenotypes, with the frequency of these cells correlating with younger age and a higher survival rate100. Also, the presence of SARS-CoV-2-specific memory CD4+ T cells and CD8+ T cells in convalescent patients correlated with mild disease99. SARS-CoV-2-specific T cells were present in individuals who were in close contact with patients with COVID-19 (refs101,102). These individuals were asymptomatic and had no evidence of infection as they tested negative for SARS-CoV-2 by quantitative reverse transcription–PCR testing and did not seroconvert101,102. This is an intriguing observation as it is unclear how individuals without confirmed infection developed SARS-CoV-2-specific T cell responses. It is possible that these individuals were transiently infected with SARS-CoV-2, rapidly cleared the infection and still developed a T cell response. However, the current evidence does not preclude the possibility that these individuals were never infected with SARS-CoV-2 and that these SARS-CoV-2-specific T cells were pre-existing common cold coronavirus-specific T cells. Further studies are warranted to characterize the SARS-CoV-2-specific T cells present in these individuals.
Pathogenic roles of T cells in coronavirus infection
Despite several lines of evidence suggesting a protective role of the T cell response in coronavirus infections, T cell-mediated pathogenesis has also been reported. This is most notable in mice infected with a murine coronavirus, mouse hepatitis virus (MHV). Infection of susceptible A/J and C3H/HeJ mice with a pneumotropic strain of MHV (MHV-1) resulted in significant pulmonary pathology. Antibody-mediated depletion of CD4+ T cells and CD8+ T cells decreased pathological changes in the lungs, suggesting that CD4+ T cells and CD8+ T cells induced pulmonary pathogenesis in these mice103. In another example, infection of the central nervous system with the neurotropic JHM strain of MHV (MHV-JHM) induced encephalitis with both acute and chronic demyelination of the central nervous system during T cell-mediated virus clearance104,105,106, with MHV-JHM-specific T cells required for both virus clearance and immunopathology. Furthermore, in patients with COVID-19, Zhou et al. identified a subset of CD4+ T cells with T helper 1-like properties that secreted high levels of granulocyte–monocyte colony-stimulating factor (GM-CSF), IL-6 and IFNγ. The frequency of these cells correlated with disease severity29. It was proposed that the cytokines secreted by this T cell subset resulted in increased levels of CD14+CD16+ inflammatory monocytes with abundant IL-6 expression that potentially led to a hyperinflammatory state in patients29. However, one caveat is that only a small number of patients were involved in that study; thus, a pathological role for this T cell subset needs to be validated in larger numbers of patients.
In another study, in which the transcriptomic profile of CD8+ T cells was characterized, a higher frequency of CD8+ T cells expressing cytotoxic effector molecules (namely, granzyme B, granzyme H, granulysin and FAS ligand) and inflammatory cytokines/chemokines (such as CCL3, CCL4, CSF2, TNF, LTA and LTB) but expressing lower levels of molecules associated with T cell exhaustion (such as TIM3, LAG3 and CD38) was observed in patients with severe disease than in patients with mild disease. Also, non-exhausted CD8+ T cells were associated with a more robust memory phenotype in patients with severe disease as evident by enriched expression of genes involving co-stimulation, prosurvival signalling and anti-apoptotic pathways107. This possibly suggests that a more robust CD8+ T cell response plays a role in mediating severe disease in patients. However, a counterargument was proposed by a separate study, where increasing expression of the T cell exhaustion markers PD1 and TIM3 was found as disease progressed from the pre-symptomatic stage to the symptomatic stage in some patients108. These results seem to contradict one another; however, the different conclusions related to the role of T cells in disease progression may reflect heterogeneity of virus-specific T cells. In this age of single-cell RNA sequencing, it is not surprising to identify different populations of cells among the same subset that perform different functions29,107,108,109. An important future goal will be to dissect the heterogeneity of virus-specific T cells so as to assess whether T cells are protective or detrimental. Also, significant changes in the frequencies of the different populations of T cells may not be linked to substantial changes in the actual number of cells due to the presence of lymphopenia, which is associated with severe disease. In addition, other studies have suggested that dysfunctional myeloid cells may contribute to a suboptimal T cell response100.
Humoral immunity and immunopathology in coronavirus infections
Coronavirus infections induce neutralizing and non-neutralizing antibodies110,111. Neutralizing antibodies target primarily the S protein by sterically blocking the interaction between the S protein and the cell surface entry receptor it binds, thereby limiting viral entry into susceptible cells. The functions of the non-neutralizing antibodies generated against coronaviruses are less clear. One possible function of non-neutralizing antibodies could involve antibody-dependent cellular cytotoxicity112. The kinetics of the SARS-CoV-2-specific antibody response differs in patients with severe disease versus patients with mild disease. High numbers of plasmablasts are detected in patients with severe disease, reflecting high viral loads113, However, in general, rapid induction of antibodies directed against SARS-CoV-2 and related coronaviruses in patients with acute disease correlates with reduced disease severity and viral load110,114,115. In convalescent patients who survived severe disease, levels of virus-specific antibodies are higher than in patients with mild disease. Induction of T cell responses with a lack of seroconversion was observed in some mild or asymptomatic cases of MERS and COVID-19, suggesting discordant T cell and antibody responses79,116. Generally, seroconversion occurs 7–14 days after symptom onset, with peak levels of antibodies attained by 15–30 days after symptom onset110,115,117. Virus-specific IgM is first detected, but then the levels of specific IgM show rapid waning110,115,117. Detectable levels of SARS-CoV-2-specific IgG with specificities for different viral proteins were seen 3–4 months after symptom onset. Levels of antibodies to SARS-CoV-2 and related coronaviruses waned over time, especially in patients with mild disease110,115,117,118. Despite waning antibody levels in the blood, SARS-CoV-2-specific memory B cells were found to increase in abundance for up to 6 months after infection77. More importantly, accumulating somatic hypermutation was observed in a temporal analysis of the antibody sequences from convalescent patients, suggesting that maturation of antibodies occurred during the convalescent phase77. Furthermore, a stable population of SARS-CoV-2-specific long-lived plasma cells was detected in the bone marrow of convalescent patients119.
SARS-CoV-2 variants escape neutralizing antibody response
During the course of the COVID-19 pandemic, several SARS-CoV-2 variants have arisen. While mutations are detected in many viral proteins, the S protein is a hotspot for mutations that enhance transmission and evade the neutralizing antibody response120,121. These variants were generally initially selected for increased transmissibility122,123. Some of the mutations in these variants mediated partial resistance to neutralizing antibodies induced by natural infection or vaccination and render some monoclonal antibodies ineffective. In the presence of a highly immunized population, these mutations may be additionally selected because they are immunoevasive120,121,124. One SARS-CoV-2 variant (the β-variant, also known as B.1.351) encodes several mutations in the S protein that enhance transmission and make this variant more resistant to neutralization by convalescent plasma and vaccine-induced sera120,121,124. In particular, the E484K mutation found in the S protein of B.1.351 is the major driver in neutralizing antibody resistance. L452R and N501Y mutations in the S protein have been shown to result in enhanced binding of SARS-CoV-2 to its entry receptor on human cells, ACE2 (refs123,125). The emergence of SARS-CoV-2 variants with increased resistance to antibody neutralization is alarming. However, correlates of protection have not been well defined, and some vaccines induce an antibody response that remains protective upon exposure to variant viruses120. Of note, the duration of immunity to original strains or variants remains unknown. It is therefore important to achieve high levels of neutralizing antibodies through vaccination to maximize the protective margin against these variants. Also, active somatic hypermutation for antibody maturation over time could enhance antibody neutralization, which could potentially offer sufficient protection against these variants77.
Potential pathogenic role of antibody response in coronavirus infection
Antibody-dependent enhancement is one of the most concerning outcomes of infection or reinfection with SARS-CoV-2 after vaccination or natural infection. However, in the case of coronaviruses, antibody-dependent enhancement, or enhanced infection of macrophages, has been observed only in felines infected with feline infectious peritonitis virus. Macrophages are productively infected with feline infectious peritonitis virus, and infection is enhanced by treatment with sera from infected cats or by vaccination with S protein-containing constructs126. Classical antibody-dependent enhancement requires interaction between virus–antibody immune complexes and Fc receptors on macrophages to enhance infection127. In SARS-CoV-2, macrophages are only abortively infected despite induction of an inflammatory response in these cells128,129; thus, classical antibody-dependent enhancement is unlikely to occur in SARS-CoV-2 infection130. In addition, in the case of SARS-CoV infection, anti-S protein antibodies have been shown to change the phenotype of lung-infiltrating macrophages from wound healing to pro-inflammatory, although this did not affect clinical disease observed after challenge131.
Another factor in antibody protection versus pathogenicity is related to Fc effector function. For example, afucosylation of the Fc region of antibodies enhances their activity by increasing Fc engagement with Fcγ receptor IIa and Fcγ receptor III (ref.132). This may result in increased protective efficacy but may also result in enhanced macrophage activation and aberrant induction of inflammatory cytokines (such as IL-6 and TNF) that are key for COVID-19 pathogenesis133,134. Macrophages that are robustly activated by afucosylated antibodies may damage pulmonary endothelial walls and induce thrombosis in the microvasculature133. High levels of afucosylated SARS-CoV-2 antibodies have been found in patients with severe COVID-19 (refs133,134,135). These results illustrate the importance of appropriate Fc modifications to prevent immunopathology mediated by antibody response.
Concluding remarks
In this Perspective, we have provided an overview of the current knowledge of innate and adaptive immune dysregulation mediated by the host (immunopathogenic) and the virus (active interference by virus). COVID-19 serves as an excellent example of the complexity and interdependent nature of host immune homeostasis. Once disrupted, as in the case of COVID-19, the consequences are a series of immunopathological changes that lead to disease progression. A more comprehensive understanding of these changes could allow us to develop more effective therapies for the treatment of patients who develop severe disease as a consequence of coronavirus infection.
Change history
17 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41577-021-00673-1
References
CDC. SARS. Basics Factsheet. https://www.cdc.gov/sars/about/fs-sars.html (2017).
WHO. EMRO. MERS outbreaks. MERS-CoV. Health topics. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (2021).
WHO. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int. (2021).
Schoggins, J. W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 6, 567–584 (2019).
Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, 6515 (2020).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, 6515 (2020).
Huang, Y.-Q. et al. No statistically apparent difference in antiviral effectiveness observed among ribavirin plus interferon-alpha, lopinavir/ritonavir plus interferon-alpha, and ribavirin plus lopinavir/ritonavir plus interferon-alpha in patients with mild to moderate coronavirus disease 2019: results of a randomized, open-labeled prospective study. Front. Pharmacol. 11, 1071 (2020).
Omrani, A. S. et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 14, 1090–1095 (2014).
Loutfy, M. R. et al. Interferon Alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290, 3222 (2003).
Shalhoub, S. Interferon beta-1b for COVID-19. Lancet 395, 1670–1671 (2020).
Consortium, W. S. T. Repurposed antiviral drugs for covid-19 — interim WHO solidarity trial results. N. Engl. J. Med. 384, 497–511 (2020).
Cameron, M. J. et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81, 8692–8706 (2007).
Lucase, C. et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020).
Galani, I. et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 22, 32–40 (2021).
Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020).
Roberts, A. et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007).
Gu, H. et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020).
Li, K. et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl Acad. Sci. USA 114, E3119–E3128 (2017).
Channappanavar, R. et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-Infected mice. Cell Host Microbe 19, 181–193 (2016).
Mahlakõiv, T. et al. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 93, 2601–2605 (2012).
Frieman, M. B. et al. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6, e1000849 (2010).
Channappanavar, R. et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 129, 3625–3639 (2019).
Li, M. et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 6, eabi9007 (2021).
Mao, T. et al. A stem-loop RNA RIG-I agonist confers prophylactic and therapeutic protection against acute and chronic SARS-CoV-2 infection in mice. Preprint at bioRxiv https://doi.org/10.1101/2021.06.16.448754 (2021).
Leist, S. R. et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020).
Bessière, P. et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog. 17, e1009427 (2021).
Tian, S. et al. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 15, 700–704 (2020).
Xu, Z. et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 8, 420–422 (2020).
Zhou, Y. et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci. Rev. 7, 998–1002 (2020).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Zhou, Z. et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27, 883–890.e2 (2020).
Israelow, B. et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 217, e20201241 (2020).
Lee, J. S. et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 5, abd1554 (2020).
Major, J. et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369, 712–717 (2020).
Sposito, B. et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell 184, 4953–4968.e16 (2021).
Broggi, A. et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 369, 706–712 (2020).
de Wilde, A. H. et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 94, 1749–1760 (2013).
Miorin, L. et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl Acad. Sci. USA 117, 28344–28354 (2020).
Katsura, H. et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27, 890–904.e8 (2020).
Lokugamage, K. G. et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94, e01410–e01420 (2020).
Xia, H. et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020).
Yuen, C.-K. et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 9, 1418–1428 (2020).
Lei, X. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810 (2020).
Frieman, M. et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J. Virol. 81, 9812–9824 (2007).
Jiang, H. et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 17, 998–1000 (2020).
Krafcikova, P., Silhan, J., Nencka, R. & Boura, E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 11, 3717 (2020).
Wang, S. et al. Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 23, 718–732 (2021).
Chen, L. et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 9, 7204–7218 (2017).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Mathew, D. et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511 (2020).
Nienhold, R. et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 11, 5086 (2020).
Barnes, B. J. et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 217, e20200652 (2020).
Giamarellos-Bourboulis, E. J. et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000.e3 (2020).
Jakubzick, C. V., Randolph, G. J. & Henson, P. M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17, 349–362 (2017).
Carter, M. J. et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 26, 1701–1707 (2020).
Ferreira, A. C. et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 7, 1–12 (2021).
Rodrigues, T. S. et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 218, e20201707 (2020).
Swanson, K. V., Deng, M. & Ting, J. P.-Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Nieto-Torres, J. L. et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485, 330–339 (2015).
Siu, K. L. et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 33, 8865–8877 (2019).
Shi, C. S., Nabar, N. R., Huang, N.-N. & Kehrl, J. H. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 5, 1–12 (2019).
DeDiego, M. L. et al. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 88, 913–924 (2014).
Wu, Y. et al. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-κB hyper-activation and inflammation. Signal. Transduct. Target. Ther. 6, 167 (2021).
Su, C. M., Wang, L. & Yoo, D. Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 11, 13464 (2021).
Cugno, M. et al. Complement activation and endothelial perturbation parallel COVID-19 severity and activity. J. Autoimmun. 116, 102560 (2021).
Gralinski, L. E. et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 9, e01753-18 (2018).
Jiang, Y. et al. Blockade of the C5a–C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg. Microbes Infect. 7, 77 (2018).
Java, A. et al. The complement system in COVID-19: friend and foe? JCI Insight 5, 15 (2020).
Magro, C. et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 220, 1–13 (2020).
Holter, J. C. et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl Acad. Sci. USA 117, 25018–25025 (2020).
Cugno, M. et al. Complement activation in patients with COVID-19: a novel therapeutic target. J. Allergy Clin. Immunol. 146, 215–217 (2020).
Zuo, Y. et al. Neutrophil extracellular traps in COVID-19. JCI Insight 5, 11 (2020).
Markiewski, M. M. & Lambris, J. D. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am. J. Pathol. 171, 715–727 (2007).
de Bont, C. M., Boelens, W. C. & Pruijn, G. J. M. NETosis, complement, and coagulation: a triangular relationship. Cell. Mol. Immunol. 16, 19–27 (2019).
Le Bert, N. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020).
Goel, R. R. et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 6, eabi6950 (2021).
Sokal, A. et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell 184, 1201–1213.e14 (2021).
Zhao, J. et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44, 1379–1391 (2016).
Mok, C. K. P. et al. T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: an observational cohort study. Lancet Infect. Dis. 21, 385–395 (2021).
Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501 (2020).
Zhao, J. et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl Acad. Sci. USA 111, 4970–4975 (2014).
Israelow, B. et al. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 6, eabl4509.
Channappanavar, R., Fett, C., Zhao, J., Meyerholz, D. K. & Perlman, S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 88, 11034–11044 (2014).
Sun, J. et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743.e5 (2020).
Chen, J. et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 84, 1289–1301 (2010).
Gu, J. & Korteweg, C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 170, 1136–1147 (2007).
de Wit, E. et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl. Acad. Sci. USA 110, 16598–16603 (2013).
Murphy, K. SARS CoV-2 detection from upper and lower respiratory tract specimens: diagnostic and infection control implications. Chest 158, 1804–1805 (2020).
Channappanavar, R., Zhao, J. & Perlman, S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 59, 118–128 (2014).
Zhao, J., Zhao, J., Rooijen, N. V. & Perlman, S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 5, e1000636 (2009).
Zhao, J., Zhao, J., Legge, K. & Perlman, S. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 121, 4921–4930 (2011).
Vijay, R. et al. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J. Exp. Med. 212, 1851–1868 (2015).
Wong, L.-Y. R. et al. Eicosanoid signaling as a therapeutic target in middle-aged mice with severe COVID-19. Preprint at bioRxiv https://doi.org/10.1101/2021.04.20.440676 (2021).
Davies, N. G. et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 26, 1205–1211 (2020).
O’Driscoll, M. et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 590, 140–145 (2021).
Le Bert, N. et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 218, e20202617 (2021).
Schulien, I. et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 27, 78–85 (2021).
Tan, A. T. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 34, 108728 (2021).
Rydyznski Moderbacher, C. et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012.e19 (2020).
Szabo, P. A. et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity 54, 797–814.e6 (2021).
Sekine, T. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183, 158–168.e14 (2020).
Wang, Z. et al. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat. Commun. 12, 1724 (2021).
Khanolkar, A. et al. Protective and pathologic roles of the immune response to mouse hepatitis virus type 1: implications for severe acute respiratory syndrome. J. Virol. 83, 9258–9272 (2009).
Lampert, P. W., Sims, J. K. & Kniazeff, A. J. Mechanism of demyelination in JHM virus encephalomyelitis. Acta Neuropathol. 24, 76–85 (1973).
Knobler, R. L., Haspel, M. V. & Oldstone, M. B. Mouse hepatitis virus type 4 (JHM strains). induced fatal central nervous system disease. I. genetic control and murine neuron as the susceptible site of disease. J. Exp. Med. 153, 832–843 (1981).
Wu, G. F., Dandekar, A. A., Pewe, L. & Perlman, S. CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J. Immunol. 165, 2278–2286 (2000).
Kusnadi, A. et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8+ T cells. Sci. Immunol. 6, abe4782 (2021).
Diao, B. et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front. Immunol. 11, 827 (2020).
Zheng, M. et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 17, 533–535 (2020).
Wang, Y. et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 130, 5235–5244 (2020).
Dogan, M. et al. SARS-CoV-2 specific antibody and neutralization assays reveal the wide range of the humoral immune response to virus. Commun. Biol. 4, 1–13 (2021).
Loos, C. et al. Evolution of early SARS-CoV-2 and cross-coronavirus immunity. mSphere 5, e00622-20 (2020).
Kuri-Cervantes, L. et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 5, eabd7114 (2020).
Zhao, J. et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T cell responses. Sci. Immunol. 2, eaan5393 (2017).
Sun, B. et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 9, 940–948 (2020).
Reynolds, C. J. et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 5, abf3698 (2020).
Jiang, X.-L. et al. Lasting antibody and T cell responses to SARS-CoV-2 in COVID-19 patients three months after infection. Nat. Commun. 12, 897 (2021).
Bilich, T. et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci. Transl. Med. 13, abf7517 (2021).
Turner, J. S. et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421–425 (2021).
Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021).
Hoffmann, M. et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393.e12 (2021).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020).
Ramanathan, M., Ferguson, I. D., Miao, W. & Khavari, P. A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 21, 1070 (2021).
Liu, Y. et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 384, 1466–1468 (2021).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020).
Takano, T. et al. Antibody-dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. Arch. Virol. 162, 3339–3345 (2017).
Wen, J. et al. Antibody-dependent enhancement of coronavirus. Int. J. Infect. Dis. 100, 483–489 (2020).
Zheng, J. et al. Severe acute respiratory syndrome coronavirus 2–induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 223, 785–795 (2021).
Hui, K. P. Y. et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 8, 687–695 (2020).
Li, D. et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 184, 4203–4219.e32 (2021).
Liu, L. et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4, e123158 (2019).
Schäfer, A. et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 218, e20201993 (2020).
Hoepel, W. et al. High titers and low fucosylation of early human anti–SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med. 13, eabf8654 (2021).
Chakraborty, S. et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 22, 67–73 (2021).
Larsen, M. D. et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science 371, eabc8378 (2021).
Bhatt, P. R. et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313 (2021).
Acknowledgements
The authors thank A. Sariol for critical review of the manuscript. This work was supported in part by grants from the US National Institutes of Health to S.P. (P01 AI060699 and R01 AI129269) and L.-Y.R.W. (T32 AI007511).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thanks A. Bertoletti, L. Enjaunes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wong, LY., Perlman, S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses — are we our own worst enemy?. Nat Rev Immunol 22, 47–56 (2022). https://doi.org/10.1038/s41577-021-00656-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-021-00656-2
This article is cited by
-
Host-directed immunotherapy of viral and bacterial infections: past, present and future
Nature Reviews Immunology (2023)
-
Comparison of infection and human immune responses of two SARS-CoV-2 strains in a humanized hACE2 NIKO mouse model
Scientific Reports (2023)
-
Interfer-on time: lessons from genetically diverse mouse models of SARS-CoV-2 infection
Genes & Immunity (2023)
-
A multicentre study reveals dysbiosis in the microbial co-infection and antimicrobial resistance gene profile in the nasopharynx of COVID-19 patients
Scientific Reports (2023)
-
Metabolic dysregulation impairs lymphocyte function during severe SARS-CoV-2 infection
Communications Biology (2023)