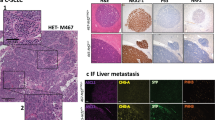
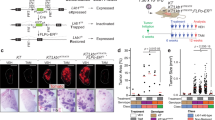
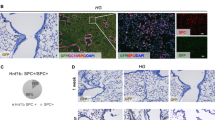
Abstract
Small cell lung cancer (SCLC) is an aggressive neuroendocrine cancer characterized by loss of function TP53 and RB1 mutations in addition to mutations in other oncogenes including MYC. Overexpression of MYC together with Trp53 and Rb1 loss in pulmonary neuroendocrine cells of the mouse lung drives an aggressive neuroendocrine low variant subtype of SCLC. However, the transforming potential of MYC amplification alone on airway epithelium is unclear. Therefore, we selectively and conditionally overexpressed MYC stochastically throughout the airway or specifically in neuroendocrine, club, or alveolar type II cells in the adult mouse lung. We observed that MYC overexpression induced carcinoma in situ which did not progress to invasive disease. The formation of adenoma or SCLC carcinoma in situ was dependent on the cell of origin. In contrast, MYC overexpression combined with conditional deletion of both Trp53 and Rb1 exclusively gave rise to SCLC, irrespective of the cell lineage of origin. However, cell of origin influenced disease latency, metastatic potential, and the transcriptional profile of the SCLC phenotype. Together this reveals that MYC overexpression alone provides a proliferative advantage but when combined with deletion of Trp53 and Rb1 it facilitates the formation of aggressive SCLC from multiple cell lineages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol: Off J Am Soc Clin Oncol. 2006;24:4539–44.
Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res. 2018;7:69–79.
George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t
Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t
Voortman J, Lee JH, Killian JK, Suuriniemi M, Wang Y, Lucchi M, et al. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci USA. 2010;107:13040–5.
Johnson BE, Russell E, Simmons AM, Phelps R, Steinberg SM, Ihde DC, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl. 1996;24:210–7.
Alves Rde C, Meurer RT, Roehe AV. MYC amplification is associated with poor survival in small cell lung cancer: a chromogenic in situ hybridization study. J cancer Res Clin Oncol. 2014;140:2021–5.
Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer cell. 2017;31:270–285.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97.
Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer cell. 2020;38:60–78.e12.
Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer cell. 2011;19:754–64. Research Support, Non-U.S. Gov’t
Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100.
Yang D, Denny SK, Greenside PG, Chaikovsky AC, Brady JJ, Ouadah Y, et al. Intertumoral heterogeneity in SCLC is influenced by the cell type of origin. Cancer discov. 2018.
Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–81.
Simpson DS, Mason-Richie NA, Gettler CA, Wikenheiser-Brokamp KA. Retinoblastoma family proteins have distinct functions in pulmonary epithelial cells in vivo critical for suppressing cell growth and tumorigenesis. Cancer Res. 2009;69:8733.
Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Dev (Camb, Engl). 2004;131:4299.
Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis. 1982;126:332–7.
Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of clara cells in normal human airway epithelium. Am J Respir Crit Care Med. 1999;159:1585–91.
Grunblatt E, Wu N, Zhang H, Liu X, Norton JP, Ohol Y, et al. MYCN drives chemoresistance in small cell lung cancer while USP7 inhibition can restore chemosensitivity. Genes Dev. 2020;34:1210–26.
Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–42.
Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, et al. Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell Rep. 2016;16:631–43.
Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res. 2018;7:32–49.
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Morton JP, Sansom OJ. MYC-y mice: from tumour initiation to therapeutic targeting of endogenous MYC. Mol Oncol. 2013;7:248–58.
Geick A, Redecker P, Ehrhardt A, Klocke R, Paul D, Halter R. Uteroglobin promoter-targeted c-MYC expression in transgenic mice cause hyperplasia of Clara cells and malignant transformation of T-lymphoblasts and tubular epithelial cells. Transgenic Res. 2001;10:501–11.
Ehrhardt A, Bartels T, Geick A, Klocke R, Paul D, Halter R. Development of pulmonary bronchiolo-alveolar adenocarcinomas in transgenic mice overexpressing murine c-myc and epidermal growth factor in alveolar type II pneumocytes. Br J cancer. 2001;84:813–8.
Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 2018;32:915–28.
Olsen RR, Ireland AS, Kastner DW, Groves SM, Spainhower KB, Pozo K, et al. ASCL1 represses a SOX9(+) neural crest stem-like state in small cell lung cancer. Genes Dev. 2021;35:847–69.
Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer cell. 2019;36:385–401. e388
Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc cooperates with ras by programming inflammation and immune suppression. Cell. 2017;171:1301–15. e1314
Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220–9.
Foroutan M, Bhuva DD, Lyu R, Horan K, Cursons J, Davis MJ. Single sample scoring of molecular phenotypes. BMC Bioinforma. 2018;19:404.
Acknowledgements
The authors thank Dr. Kate Sutherland for Ad5-Cre viruses and helpful discussions. We thank the Monash Health Translational Precinct Histology Platform, Monash MicroImaging, Monash Medical Center Animal Facility, Monash Health Translational Precinct Flow Cytometry Core, Monash Biomedical Imaging, Beijing Genomics Institute (BGI) and the Monash Bioinformatics Platform for providing core and technical services.
Funding
This work was supported by the Operational Infrastructure Support Program by the Victorian Government of Australia. D.J. Gough is supported by a Victorian Cancer Agency Mid-Career Fellowship (MCRF19033) and a grant in aid from the Cancer Council of Victoria (GNT1140528).
Author information
Authors and Affiliations
Contributions
Study conception and design DJG. Experiments and Data Analysis: all authors, Manuscript writing DJG and JC, manuscript editing all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, J., Guanizo, A., Luong, Q. et al. Lineage-restricted neoplasia driven by Myc defaults to small cell lung cancer when combined with loss of p53 and Rb in the airway epithelium. Oncogene 41, 138–145 (2022). https://doi.org/10.1038/s41388-021-02070-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02070-3
This article is cited by
-
Genomic and transcriptomic profiling of combined small-cell lung cancer through microdissection: unveiling the transformational pathway of mixed subtype
Journal of Translational Medicine (2024)
-
Genetically-engineered mouse models of small cell lung cancer: the next generation
Oncogene (2024)
-
MYC drives platinum resistant SCLC that is overcome by the dual PI3K-HDAC inhibitor fimepinostat
Journal of Experimental & Clinical Cancer Research (2023)
-
Data-driven structural analysis of small cell lung cancer transcription factor network suggests potential subtype regulators and transition pathways
npj Systems Biology and Applications (2023)
-
Novel Therapeutic Options for Small Cell Lung Cancer
Current Oncology Reports (2023)