Abstract
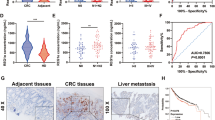
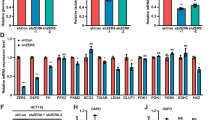
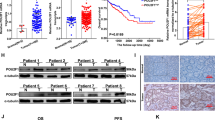
Tumor metabolic reprogramming ensures that cancerous cells obtain sufficient building blocks, energy, and antioxidants to sustain rapid growth and for coping with oxidative stress. Neurogenic differentiation factor 1 (NeuroD1) is upregulated in various types of tumors; however, its involvement in tumor cell metabolic reprogramming remains unclear. In this study, we report that NeuroD1 is positively correlated with glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme in the pentose phosphate pathway (PPP), in colorectal cancer cells. In addition, the regulation of G6PD by NeuroD1 alters tumor cell metabolism by stimulating the PPP, leading to enhanced production of nucleotides and NADPH. These, in turn, promote DNA and lipid biosynthesis in tumor cells, while decreasing intracellular levels of reactive oxygen species. Mechanistically, we showed that NeuroD1 binds directly to the G6PD promoter to activate G6PD transcription. Consequently, tumor cell proliferation and colony formation are enhanced, leading to increased tumorigenic potential in vitro and in vivo. These findings reveal a novel function of NeuroD1 as a regulator of G6PD, whereby its oncogenic activity is linked to tumor cell metabolic reprogramming and regulation of the PPP. Furthermore, NeuroD1 represents a potential target for metabolism-based anti-tumor therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Warburg O. On the origin of cancer cells. Science. 1956;123:309–14.
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20.
DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43.
Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82.
Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–6.
Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–44.
Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–28.
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3.
Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, et al. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. Embo j. 2016;35:24–45.
Kim JW, Seghers V, Cho JH, Kang Y, Kim S, Ryu Y, et al. Transactivation of the mouse sulfonylurea receptor I gene by BETA2/NeuroD. Mol Endocrinol. 2002;16:1097–107.
Marsich E, Vetere A, Di Piazza M, Tell G, Paoletti S. The PAX6 gene is activated by the basic helix-loop-helix transcription factor NeuroD/BETA2. Biochem J. 2003;376:707–15.
Martin CC, Svitek CA, Oeser JK, Henderson E, Stein R, O’Brien RM. Upstream stimulatory factor (USF) and neurogenic differentiation/beta-cell E box transactivator 2 (NeuroD/BETA2) contribute to islet-specific glucose-6-phosphatase catalytic-subunit-related protein (IGRP) gene expression. Biochem J. 2003;371:675–86.
Moates JM, Nanda S, Cissell MA, Tsai MJ, Stein R. BETA2 activates transcription from the upstream glucokinase gene promoter in islet beta-cells and gut endocrine cells. Diabetes. 2003;52:403–8.
Enya M, Horikawa Y, Kuroda E, Yonemaru K, Tonooka N, Tomura H, et al. Mutations in the small heterodimer partner gene increase morbidity risk in Japanese type 2 diabetes patients. Hum Mutat. 2008;29:E271–277.
Huang P, Kishida S, Cao D, Murakami-Tonami Y, Mu P, Nakaguro M, et al. The neuronal differentiation factor NeuroD1 downregulates the neuronal repellent factor Slit2 expression and promotes cell motility and tumor formation of neuroblastoma. Cancer Res. 2011;71:2938–48.
Lu F, Kishida S, Mu P, Huang P, Cao D, Tsubota S, et al. NeuroD1 promotes neuroblastoma cell growth by inducing the expression of ALK. Cancer Sci. 2015;106:390–6.
Boulay G, Awad ME, Riggi N, Archer TC, Iyer S, Boonseng WE, et al. OTX2 activity at distal regulatory elements shapes the chromatin landscape of group 3 medulloblastoma. Cancer Disco. 2017;7:288–301.
Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–.e347.
Wang Y, Su DW, Gao L, Ding GL, Ni CR, Zhu MH. Effect of NeuroD gene silencing on the migration and invasion of human pancreatic cancer cells PANC-1. Cell Biochem Biophys. 2014;69:487–94.
Osborne JK, Guerra ML, Gonzales JX, McMillan EA, Minna JD, Cobb MH. NeuroD1 mediates nicotine-induced migration and invasion via regulation of the nicotinic acetylcholine receptor subunits in a subset of neural and neuroendocrine carcinomas. Mol Biol Cell. 2014;25:1782–92.
Osborne JK, Larsen JE, Shields MD, Gonzales JX, Shames DS, Sato M, et al. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci USA. 2013;110:6524–9.
Lei K, Li W, Huang C, Li Y, Alfason L, Zhao H, et al. Neurogenic differentiation factor 1 promotes colorectal cancer cell proliferation and tumorigenesis by suppressing the p53/p21 axis. Cancer Sci. 2020;111:175–85.
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115.
Xu H, Liu L, Li W, Zou D, Yu J, Wang L, et al. Transcription factors in colorectal cancer: molecular mechanism and therapeutic implications. Oncogene. 2021;40:1555–69.
Reina-Campos M, Diaz-Meco MT, Moscat J. The dual roles of the atypical protein kinase Cs in cancer. Cancer Cell. 2019;36:218–35.
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10:3499.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308.
Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–69.
Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–54.
Yang HC, Wu YH, Yen WC, Liu HY, Hwang TL, Stern A, et al. The Redox Role of G6PD in Cell Growth, Cell Death, and Cancer. Cells. 2019;8:1055.
Wang J, Sui Y, Li Q, Zhao Y, Dong X, Yang J, et al. Effective inhibition of MYC-amplified group 3 medulloblastoma by FACT-targeted curaxin drug CBL0137. Cell Death Dis. 2020;11:1029.
Cheng Y, Liao S, Xu G, Hu J, Guo D, Du F, et al. NeuroD1 dictates tumor cell differentiation in medulloblastoma. Cell Rep. 2020;31:107782.
Owonikoko TK, Dwivedi B, Chen Z, Zhang C, Barwick B, Ernani V, et al. YAP1 expression in SCLC defines a distinct subtype with T-cell-inflamed phenotype. J Thorac Oncol. 2021;16:464–76.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97.
Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–72.
Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38:60–78.e12.
Cindolo L, Franco R, Cantile M, Schiavo G, Liguori G, Chiodini P, et al. NeuroD1 expression in human prostate cancer: can it contribute to neuroendocrine differentiation comprehension? Eur Urol. 2007;52:1365–73.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Wang M, Ji S, Shao G, Zhang J, Zhao K, Wang Z, et al. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin Transl Oncol. 2018;20:906–11.
Ma X, Wang L, Huang D, Li Y, Yang D, Li T, et al. Polo-like kinase 1 coordinates biosynthesis during cell cycle progression by directly activating pentose phosphate pathway. Nat Commun. 2017;8:1506.
Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–15.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative Stress in Cancer. Cancer Cell. 2020;38:167–97.
Cheng J, Huang Y, Zhang X, Yu Y, Wu S, Jiao J, et al. TRIM21 and PHLDA3 negatively regulate the crosstalk between the PI3K/AKT pathway and PPP metabolism. Nat Commun. 2020;11:1880.
Shan C, Lu Z, Li Z, Sheng H, Fan J, Qi Q, et al. 4-hydroxyphenylpyruvate dioxygenase promotes lung cancer growth via pentose phosphate pathway (PPP) flux mediated by LKB1-AMPK/HDAC10/G6PD axis. Cell Death Dis. 2019;10:525.
Liu CL, Hsu YC, Lee JJ, Chen MJ, Lin CH, Huang SY, et al. Targeting the pentose phosphate pathway increases reactive oxygen species and induces apoptosis in thyroid cancer cells. Mol Cell Endocrinol. 2020;499:110595.
Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–.e1287.
Liu Y, Hou X, Liu M, Yang Z, Bi Y, Zou H, et al. XBP1 silencing decreases glioma cell viability and glycolysis possibly by inhibiting HK2 expression. J Neurooncol. 2016;126:455–62.
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–.e157.
Giles ED, Wellberg EA, Astling DP, Anderson SM, Thor AD, Jindal S, et al. Obesity and overfeeding affecting both tumor and systemic metabolism activates the progesterone receptor to contribute to postmenopausal breast cancer. Cancer Res. 2012;72:6490–501.
Ku HC, Cheng CF. Master regulator activating transcription factor 3 (ATF3) in metabolic homeostasis and cancer. Front Endocrinol (Lausanne). 2020;11:556.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Park AK, Lee JY, Cheong H, Ramaswamy V, Park SH, Kool M, et al. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer. 2019;19:571.
Santana-Codina N, Roeth AA, Zhang Y, Yang A, Mashadova O, Asara JM, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat Commun. 2018;9:4945.
Almeida AS, Soares NL, Sequeira CO, Pereira SA, Sonnewald U, Vieira HLA. Improvement of neuronal differentiation by carbon monoxide: Role of pentose phosphate pathway. Redox Biol. 2018;17:338–47.
Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell. 2017;32:71–87.e77.
Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao H, et al. Transcription factor YY1 promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res. 2018;78:4549–62.
Acknowledgements
We thank Dr. Bert Vogelstein (School of Medicine, Johns Hopkins University) for kindly providing wild-type and p53-null HCT116p53null cell lines. This work was supported by grants from the National Natural Science Foundation of China (11832008, 31871367, and 81872273); and the Natural Science Foundation of Chongqing (cstc2018jcyjAX0411 and cstc2018jcyjAX0374).
Author information
Authors and Affiliations
Contributions
V.K. and S.W. conceived the project, analyzed and interpreted the experimental results; V.K. designed the most experiments of the project and wrote the manuscript. Z.L., Y.H., and Y.L. performed most of the experiments. J.L. performed part of plasmid constructions and cell counting assay. G.S. analyzed part of the data and provided part of the material. H.Z. collected human clinical samples and performed clinical samples analysis. M.M. designed shRNA target sites and analyzed part of the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Z., He, Y., Li, Y. et al. NeuroD1 promotes tumor cell proliferation and tumorigenesis by directly activating the pentose phosphate pathway in colorectal carcinoma. Oncogene 40, 6736–6747 (2021). https://doi.org/10.1038/s41388-021-02063-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02063-2
This article is cited by
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
Metabolic reprogramming in colorectal cancer: regulatory networks and therapy
Cell & Bioscience (2023)
-
The p52-ZER6/G6PD axis alters aerobic glycolysis and promotes tumor progression by activating the pentose phosphate pathway
Oncogenesis (2023)
-
LINC00839 Promotes Neuroblastoma Progression by Sponging miR-454-3p to Up-Regulate NEUROD1
Neurochemical Research (2022)
-
CTP synthase: the hissing of the cellular serpent
Histochemistry and Cell Biology (2022)