Abstract
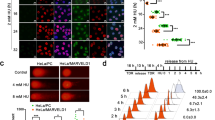
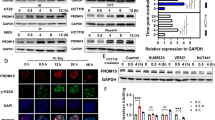
Colorectal carcinoma (CRC) is the second most deadly cancer worldwide. Therapies that take advantage of DNA repair defects have been explored in various tumors but not yet systematically in CRC. Here, we found that Diphosphoinositol Pentakisphosphate Kinase 2 (PPIP5K2), an inositol pyrophosphate kinase, was highly expressed in CRC and associated with a poor prognosis of CRC patients. In vitro and in vivo functional studies demonstrated that PPIP5K2 could promote the proliferation and migration ability of CRC cells independent of its inositol pyrophosphate kinase activity. Mechanically, S1006 dephosphorylation of PPIP5K2 could accelerate its dissociation with 14-3-3 in the cytoplasm, resulting in more nuclear distribution. Moreover, DNA damage treatments such as doxorubicin (DOX) or irradiation (IR) could induce nuclear translocation of PPIP5K2, which subsequently promoted homologous recombination (HR) repair by binding and recruiting RPA70 to the DNA damage site as a novel scaffold protein. Importantly, we verified that S1006 dephosphorylation of PPIP5K2 could significantly enhance the DNA repair ability of CRC cells through a series of DNA repair phenotype assays. In conclusion, PPIP5K2 is critical for enhancing the survival of CRC cells via facilitating DNA HR repair. Our findings revealed an unrecognized biological function and mechanism model of PPIP5K2 dependent on S1006 phosphorylation and provided a potential therapeutic target for CRC patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number as RDDB2021481511.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68:394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, et al. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–8.
Cremolini C, Schirripa M, Antoniotti C, Moretto R, Salvatore L, Masi G, et al. First-line chemotherapy for mCRC—a review and evidence-based algorithm. Nat Rev Clin Oncol. 2015;12:607–19.
Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204.
Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5.
Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64.
Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol. 2018;15:564–76.
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34.
Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60.
Chakraborty A. The inositol pyrophosphate pathway in health and diseases. Biol Rev Camb Philos Soc. 2018;93:1203–27.
Machkalyan G, Hèbert TE, Miller GJ. PPIP5K1 suppresses etoposide-triggered apoptosis. J Mol Signal. 2016;11:4.
Gu C, Nguyen HN, Ganini D, Chen Z, Jessen HJ, Gu Z, et al. KO of 5-InsP(7) kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc Natl Acad Sci USA. 2017;114:11968–73.
Chen H, Sun X, Ge W, Qian Y, Bai R, Zheng S. A seven-gene signature predicts overall survival of patients with colorectal cancer. Oncotarget. 2017;8:95054–65.
Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, et al. The target landscape of clinical kinase drugs. Science (New York, NY) 2017;358:eaan4368.
Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2011;8:111–6.
Yong ST, Nguyen HN, Choi JH, Bortner CD, Williams J, Pulloor NK, et al. Identification of a functional nuclear translocation sequence in hPPIP5K2. BMC Cell Biol. 2015;16:17.
Yaffe MB. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57.
Aghazadeh Y, Papadopoulos V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov Today. 2016;21:278–87.
Moon S, Kim W, Kim S, Kim Y, Song Y, Bilousov O, et al. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61–71.
Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–71.
Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, et al. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70:4385–93.
Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–6.
Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7:278–85.
Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37.
Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23.
Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23:9025–33.
Yang Q, Zhu Q, Lu X, Du Y, Cao L, Shen C, et al. G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc Natl Acad Sci USA. 2017;114:E6054–e6063.
Bhat KP, Cortez D. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat Struct Mol Biol. 2018;25:446–53.
Cai MY, Dunn CE, Chen W, Kochupurakkal BS, Nguyen H, Moreau LA, et al. Cooperation of the ATM and Fanconi Anemia/BRCA Pathways in Double-Strand Break End Resection. Cell Rep. 2020;30:2402–.e2405.
Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–2.
Randall TA, Gu C, Li X, Wang H, Shears SB. A two-way switch for inositol pyrophosphate signaling: evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv Biol Regul. 2020;75:100674.
Thota SG, Bhandari R. The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J Biosci. 2015;40:593–605.
Pulloor NK, Nair S, McCaffrey K, Kostic AD, Bist P, Weaver JD, et al. Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in Type-I interferon response. PLoS Pathog. 2014;10:e1003981.
Pöhlmann J, Risse C, Seidel C, Pohlmann T, Jakopec V, Walla E, et al. The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 2014;10:e1004586.
Jadav RS, Kumar D, Buwa N, Ganguli S, Thampatty SR, Balasubramanian N, et al. Deletion of inositol hexakisphosphate kinase 1 (IP6K1) reduces cell migration and invasion, conferring protection from aerodigestive tract carcinoma in mice. Cell Signal. 2016;28:1124–36.
Morrison BH, Bauer JA, Hu J, Grane RW, Ozdemir AM, Chawla-Sarkar M, et al. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21:1882–9.
Khaled ML, Bykhovskaya Y, Gu C, Liu A, Drewry MD, Chen Z, et al. PPIP5K2 and PCSK1 are candidate genetic contributors to familial keratoconus. Sci Rep. 2019;9:19406.
Yousaf R, Gu C, Ahmed ZM, Khan SN, Friedman TB, Riazuddin S, et al. Mutations in diphosphoinositol-pentakisphosphate kinase PPIP5K2 are associated with hearing loss in human and mouse. PLoS Genet. 2018;14:e1007297.
Wang H, Godage HY, Riley AM, Weaver JD, Shears SB, Potter BV. Synthetic inositol phosphate analogs reveal that PPIP5K2 has a surface-mounted substrate capture site that is a target for drug discovery. Chem Biol. 2014;21:689–99.
Pendergast AM. Stress and death: breaking up the c-Abl/14-3-3 complex in apoptosis. Nat Cell Biol. 2005;7:213–4.
Nihira K, Taira N, Miki Y, Yoshida K. TTK/Mps1 controls nuclear targeting of c-Abl by 14-3-3-coupled phosphorylation in response to oxidative stress. Oncogene. 2008;27:7285–95.
Graves PR, Lovly CM, Uy GL, Piwnica-Worms H. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene. 2001;20:1839–51.
Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23.
Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta. 2011;1813:1938–45.
Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106.
Gao SS, Guan H, Yan S, Hu S, Song M, Guo ZP, et al. TIP60 K430 SUMOylation attenuates its interaction with DNA-PKcs in S-phase cells: facilitating homologous recombination and emerging target for cancer therapy. Sci Adv. 2020;6:eaba7822.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China [grant number 2017YFC1309000]; the National Natural Science Foundation of China [grant numbers 81972227, 81730072, 82072608, 81872001, and 82002467]; the Natural Science Foundation of Guangdong [grant numbers 2020A151501021 and 2020A1515011020]; the Guangzhou Science and Technology Plan Projects [grant number 201904020044] and the China Postdoctoral Science Foundation (2020M672999).
Author information
Authors and Affiliations
Contributions
D.X. and F.-W.W. conceived and devised the study. C.-H.C. and F.-W.W. designed the experiments and analysis. H.L., K.H., X.-P.L., J.-H.C., J.Z., Z.-C.X., S.L., J.-L.L., J.-L.D. and J. L., performed the experiments. M.-Y.C., L.L. and J.-W.C. performed bioinformatics and statistical analysis. C.-H.C., H.L. and K.H. analyzed and interpreted the data. Y.-J.F., Z.-Z.P. and F.W. provided CRC patients tissue samples and clinical information. D.X. and F.-W.W. supervised the research and together with C.-H.C., H.L. and K.H. wrote the paper. All authors approved the submitted paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cao, CH., Ling, H., Han, K. et al. PPIP5K2 promotes colorectal carcinoma pathogenesis through facilitating DNA homologous recombination repair. Oncogene 40, 6680–6691 (2021). https://doi.org/10.1038/s41388-021-02052-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02052-5
This article is cited by
-
Functions, Mechanisms, and therapeutic applications of the inositol pyrophosphates 5PP-InsP5 and InsP8 in mammalian cells
Journal of Cardiovascular Translational Research (2024)