Abstract
Intestinal stem cell (ISC) differentiation is regulated precisely by a niche in the crypt, where lymphocytes may interact with stem and transient amplifying (TA) cells. However, whether and how lymphocyte–stem/TA cell contact affects ISC differentiation is largely unknown. Here, we uncover a novel role of T cell–stem/TA cell contact in ISC fate decisions. We show that intestinal lymphocyte depletion results in skewed ISC differentiation in mice, which can be rescued by T cell transfer. Mechanistically, integrin αEβ7 expressed on T cells binds to E-cadherin on ISCs and TA cells, triggering E-cadherin endocytosis and the consequent Wnt and Notch signaling alterations. Blocking αEβ7−E-cadherin adhesion suppresses Wnt signaling and promotes Notch signaling in ISCs and TA cells, leading to defective ISC differentiation. Thus, αEβ7+ T cells regulate ISC differentiation at single-cell level through cell–cell contact-mediated αEβ7−E-cadherin adhesion signaling, highlighting a critical role of the T cell–stem/TA cell contact in maintaining intestinal homeostasis.
Similar content being viewed by others
Introduction
The gut epithelium is the most rapidly self-renewing tissue in adult mammals and provides the first line of host defense by preventing the entry of potentially dangerous microorganisms. The daily constant self-renewal of the gut epithelium is driven by intestinal stem cells (ISCs), also known as Lgr5+ crypt base columnar (CBC) cells, that reside in the base of the crypts. The crypt bottom provides a unique niche that controls ISC behavior and numbers. Upon loss of contact to their niche, ISCs differentiate into transit-amplifying (TA) cells and then migrate upwards towards crypt-villus junction to generate mature absorptive and secretory epithelial cells.1,2 ISCs can mainly differentiate into five different cell lineages: the antibacterial peptide-secreting Paneth cells, mucus-secreting goblet cells, hormone-secreting enteroendocrine cells, chemosensory tuft cells and absorptive enterocytes.3 In the villi, enterocyte is the major population and a few goblet cells, enteroendocrine cells and tuft cells distribute at various intervals. These four types of cells form and mature during migration from the crypt to the villus tip. By contrast, Paneth cells differentiate as they move down to the base of the crypt.
ISC differentiation is precisely regulated by the external signals from the niche in the crypts.4 Multiple signaling pathways such as the Wnt/β-catenin axis, Notch signaling and TGF-β/BMP pathways are involved in the regulation of ISC–niche communications.5,6,7,8 Perturbations of these signaling pathways can impair ISC self-renew and differentiation, leading to increased susceptibility to intestinal diseases. Wnt/β-catenin signaling has a crucial role in the maintenance and proliferation of ISCs.9,10 Inhibition of Wnt pathway results in reduced epithelial proliferation and absence of secretory cell lineages.11 ISC niche cells, including the Paneth cells and the stromal cells surrounding the crypt, play an active role in maintaining Wnt signaling by secreting Wnt ligands.12,13,14,15 Notch signaling is also crucial for the maintenance and fate decisions of ISCs.6,16 Notch activation upregulates the expression of hes family bHLH transcription factor 1 (Hes1), which in turn inhibits the expression of mouse atonal homolog 1 (Math1, also known as Atoh1).17 Therefore, Notch activation drives Hes1-dependent absorptive lineage differentiation, while Notch inactivation drives Math1-dependent secretory lineage differentiation.18 Balanced activation of Wnt and Notch signaling is required for maintaining the stem cell pool, which is lost without either one of these signals.19,20 However, the more polarized Wnt and Notch activation is observed in secretory (Wnt high and Notch low) and absorptive (Wnt low and Notch high) progenitor cells,21 which determines the secretory and absorptive lineage differentiation.
Among the components of the niche, immune cells play important roles in intestinal epithelial barrier function and tissue regeneration by secreting different cytokines under pathological conditions. Gut-residing innate lymphoid cell-derived interleukin 22 (IL-22) promotes ISC-mediated epithelial regeneration after intestinal injury.22,23 T helper (Th) cell-derived pro- and anti-inflammatory cytokines modulate ISC renewal and differentiation in opposing ways during infection.24 Pro-inflammatory cytokines (interferon-γ, IL-13 and IL-17) from Th1, Th2 and Th17 cells suppress ISC renewal and promote ISC differentiation, whereas Treg-derived IL-10 promotes ISC renewal.24
Gut-homing receptor β7 integrins α4β7 and αEβ7 are responsible for efficient trafficking and retention of lymphocytes in the gut. Loss of β7 integrins leads to a reduced number of intestinal intraepithelial lymphocytes (IELs) and lamina propria lymphocytes (LPLs) in mice.25,26 A recent study shows that the ISC compartment is the primary intestinal site targeted by T cells after bone marrow transplantation. Adhesion molecule mucosal addressin cell adhesion molecule-1 (MAdCAM-1), the ligand for integrin α4β7, localizes to the crypt region vessels and directs T cells homing to the ISC compartment.27 Notably, lymphocytes often directly interact with intestinal epithelial cells (IECs), including stem and TA cells, in the epithelium and sub-epithelial space in the crypts.27,28,29,30 However, the functions of these lymphocyte–epithelial cell interactions in the crypts remain obscure.
Here, using single-cell RNA sequencing (scRNA-seq) and immunostaining, we showed that intestinal lymphocyte depletion resulted in aberrant intestinal epithelium differentiation in mice. We further characterized that T cells regulated Wnt and Notch pathways in ISCs and TA cells via adhesion signaling triggered by the interaction between αEβ7 on T cells and E-cadherin on ISCs and TA cells. Blocking this adhesion suppressed Wnt signaling and promoted Notch signaling in ISCs and TA cells, leading to defective IEC differentiation. Our study revealed a new functional axis for ISC fate decisions through integrin αEβ7−E-cadherin adhesion signaling mediated by T cell–stem/TA cell contact.
Results
Intestinal lymphocyte depletion causes aberrant IEC differentiation
To obtain a spatial overview of lymphocytes in the small intestine, we performed immunofluorescent staining using tissue sections of wild-type (WT) mice. This revealed that T and B cells were scattered throughout the crypts (Fig. 1a, b). Interestingly, about half of the T cells were found in contact with the crypt IECs directly (Fig. 1a), whereas B cell–IEC contact was barely observed (Fig. 1b). Along the crypt axis, T cells were distributed both in the stem cell compartment and TA cell compartment (Fig. 1c, d). Some T cells directly contacted with the ISCs and upper crypt epithelial cells (Fig. 1c, d; Supplementary information, Videos S1, S2).
a, b Representative immunofluorescent images of CD3+ T cells (a) and CD19+ B cells (b) in the small intestinal crypts of WT and β7−/− mice (left panel). Crypt epithelial cells were identified by E-cadherin (green); CD3+ T cells were in red; CD19+ B cells were in violet. The sections were counterstained with DAPI (blue). T cells in contact with crypt epithelial cells are highlighted with white arrows. Scale bars, 50 μm. The graphs (right panel) show the quantification of total T or B cells and T or B cells in contact with crypt epithelial cells per crypt section, and the frequency of contacted T cells within total T cells in the crypt. n = 5 mice, more than 5 fields per mouse. Data are represented as means ± SEM. ****P < 0.0001, ***P < 0.001, **P < 0.01, ns, no significant difference, t-test. c Representative immunofluorescent images of direct contacts between T cells and crypt epithelial cells in Lgr5-GFP mice. Crypt epithelial cells were stained by E-cadherin (blue) and CD3+ T cells were in red. ISCs were identified by Lgr5-GFP and the sections were counterstained with DAPI (white). Scale bars, 50 μm. d Quantification of T cell–IEC contact at each position in the small intestinal crypts of WT and β7−/− mice according to the scheme in the inset (left panel). Data were collected from 115 crypts of WT mice (n = 6) and 129 crypts of β7−/− mice (n = 5).
To investigate the potential role of lympho–epithelial interactions in the crypts, we depleted the intestinal lymphocytes by abolishing the expression of the gut-homing receptor integrins α4β7 and αEβ7 using integrin β7 knockout (β7−/−) mice (Fig. 1a, b; Supplementary information, Fig. S1a).25,31 T cell number and T cell–IEC contacts were significantly reduced in the small intestinal crypt region in β7−/− mice (Fig. 1a). Decreased T cell–IEC contacts were observed at most crypt cell positions in β7−/− mice (Fig. 1d). The length of villi and crypts at the proximal, middle and distal regions of the small intestine showed no significant changes (Supplementary information, Fig. S1b). We next crossed β7−/− mice with Lgr5-EGFP-IRES-creERT2 (Lgr5-GFP) mice. Although there were no obvious changes in the numbers of Lgr5+ ISCs (Fig. 2a) and total crypt cells (Supplementary information, Fig. S2a) in β7−/− mice, an increased EdU incorporation was observed in the crypt region (Fig. 2b), indicating the enhanced cell proliferation. Notably, immunohistochemical staining results showed decreased lysozyme (LYZ)+ Paneth cells and periodic acid-Schiff (PAS)+ goblet cells, and increased alkaline phosphatase (AP)+ enterocytes with decreased AP activity in β7−/− mice (Fig. 2c). Consistently, we observed the decreased expression of Paneth cell markers Lyz1, Defa5 and goblet cell markers Muc2, Muc5ac, and the increased expression of enterocyte markers Alpi, Anpep in the small intestine of β7−/− mice (Supplementary information, Fig. S2b). To investigate the skewed ISC differentiation in β7−/− mice at the clonal level, we traced Lgr5+ cells and their offspring using Lgr5-EGFP-IRES-creERT2; Rosa26-confetti (Lgr5-Confetti) mice with WT (control) or deficient β7 integrin (β7−/−). The control Lgr5-Confetti mice showed polyclonal crypts and villi by activating one of the Confetti colors (membranous CFP, cytoplastic YFP and RFP, nuclear GFP) in individual Lgr5+ ISCs (Fig. 2d–g; Supplementary information, Fig. S2c). In combination with immunostaining of differentiated cell markers at 10 days after a single tamoxifen (TAM) injection (dpt10), Lgr5-Confetti; β7−/− mice displayed decreased Muc2+ goblet cells and LYZ+ Paneth cells and increased Fabp1+ enterocytes and ChgA+ EECs (Fig. 2d–g; Supplementary information, Fig. S2c). In addition to using β7−/− mice, we also blocked the function of α4β7 and αEβ7 integrins in WT mice using integrin β7 blocking mAb FIB504. Consistent results were observed in these mice including significant decrease of T/B cells and T cell–epithelial cell contact in the intestinal crypts (Supplementary information, Fig. S3a, b), decreased Paneth cells and goblet cells, and increased enterocytes with decreased AP activity (Supplementary information, Fig. S3c). Collectively, the results above suggest an important role of gut-residing lymphocytes in maintaining normal IEC differentiation.
a Representative images (left) and quantification (right) of Lgr5+ ISCs per crypt section in control (Lgr5-GFP) and β7−/− (Lgr5-GFP; β7−/−) mice. The sections were counterstained with DAPI (red). Scale bars, 50 μm. n = 72 crypts from three WT mice and n = 60 crypts from three β7−/− mice. Data are represented as means ± SEM, ns, no significant difference, t-test. b Representative images (left) and quantification (right) of EdU+ (red) cells in the crypts of WT and β7−/− mice. The sections were counterstained with DAPI (green). Scale bars, 50 μm. n = 104 crypts from three WT mice and n = 100 crypts from three β7−/− mice. Data are represented as means ± SEM. ***P < 0.001, t-test. c Immunohistochemical analysis of paraffin-embedded small intestinal sections from WT and β7−/− mice. Paneth cells (top), goblet cells (middle) and enterocytes (bottom) were visualized by LYZ staining, PAS staining and AP staining, respectively (left panel). Scale bars, 50 μm. Quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus (right panel). n = 6 mice per group, more than 8 fields per mouse. Data are represented as means ± SEM, ***P < 0.001, **P < 0.01, t-test. d–g Multi-lineage tracing in control (Lgr5-Confetti) and β7−/− (Lgr5-Confetti; β7−/−) mice 10 days after one dose TAM treatment. The Lgr5-Confetti mice showed polyclonal crypts and villi by activating one of the Confetti colors (membranous CFP, cytoplastic YFP and RFP, nuclear GFP) in individual Lgr5+ ISCs. Differentiation markers were detected by immunofluorescence within Lgr5 lineage stripes, with Muc2 for goblet cells (d), LYZ for Paneth cells (e), Fabp1 for enterocytes (f) and ChgA for enteroendocrine cells (EECs) (g). Left, representative images of one Confetti color lineage for each differentiated cell type. Yellow arrows identify cells co-expressing the Confetti color and the marker of the indicated cell types. Scale bars, 50 μm. Right, percentage of the differentiated cell types in each clone. More than 100 clones were analyzed for each cell type. Data are represented as means ± SEM, ***P < 0.001, **P < 0.01, *P < 0.05, t-test.
Intestinal lymphocyte depletion alters Wnt and Notch signaling in the crypts
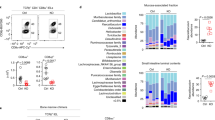
Next, we isolated the crypts and transcriptionally profiled 13,352 individual IECs from WT mice (n = 3 mice), and 10,763 IECs from β7−/− mice (n = 3 mice) using scRNA-seq. Cells were partitioned into 8 groups by unsupervised clustering and annotated by distinct cell types or states according to the known marker genes for IECs as previously described (Fig. 3a; Supplementary information, Fig. S4).32 Although the proportions of stem and TA cells showed no significant changes, both the secretory and absorptive lineages were altered in β7−/− mice, including the decreased percentage of Paneth cells and goblet cells and a marginal increase in enterocytes (Fig. 3b).
a Single cell transcriptomic analysis of 13,352 individual IECs from WT mice (n = 3) and 10,763 IECs from β7−/− mice (n = 3). Unsupervised clustering resolved at least 8 distinct types of cells, and data are presented as UMAP projection to visualize variation in single cell transcriptomes. All the cells were colored by orthogonally generated clusters labeled by manual cell type annotation. TA, transient amplifying cells; E, enterocytes; SP, secretory progenitor; EEC, enteroendocrine cells. b Cluster frequency analysis depicting the fraction of cells in WT and β7−/− mice that contributed to each cluster in panel a. n = 3 mice for each group, data are represented as means ± SEM. **P < 0.01, *P < 0.05, ·P < 0.1, likelihood ratio test. c UMAP plots of ISCs that are extracted from scRNA-seq clusters of WT and β7−/− mice in panel a. Left, UMAP plot of 2063 ISCs in WT mice and 1507 ISCs in β7−/− mice; Right, ISC subset assignment by orthogonally generated clusters with post hoc annotation. d UMAP plots of TA cells that are extracted from scRNA-seq clusters of WT and β7−/− mice in panel a. Left, UMAP plot of 2789 TA cells in WT mice and 2,097 TA cells in β7−/− mice; Right, TA cell subset assignment by orthogonally generated clusters with post hoc annotation. e Dot plots showing mean signature scores of Wnt (left) and Notch (right) pathways in the indicated ISC subsets. Fraction of each ISC subset (circle size) and the signature’s mean score (color bar) for each ISC subset (rows) in each genotype (columns) were shown. ***P < 10−15, **P < 10−10, *P < 10−5, Mann–Whitney U-test. f Dot plots showing mean signature scores of Wnt (left) and Notch (right) pathways in the indicated TA cell subsets. Fraction of each TA subset (circle size) and the signature’s mean score (color bar) for each TA subset (rows) in each genotype (columns) were shown. ***P < 10−15, **P < 10−10, *P < 10−5, Mann–Whitney U-test.
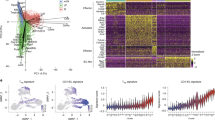
To further investigate the alternations in the ISCs and TA cells in β7−/− mice, ISCs and TA cells were further partitioned into six subsets, ISC-I, -II, -III (Fig. 3c; Supplementary information, Fig. S5a) and TA-I, -II, -III (Fig. 3d; Supplementary information Fig. S5b). Within ISC subsets, ISC-I showed the highest stemness signature and lowest cell-cycle signature, while ISC-II and -III are more differentiated and proliferative (Supplementary information, Fig. S5c, d). Similarly, within TA subsets, TA-I expressed higher stemness signature and lower cell-cycle signature than the other two subtypes (Supplementary information, Fig. S5e, f). Although no significant changes were observed in the proportion of all subsets within ISCs and TA cells in β7−/− mice, Wnt signaling was downregulated while the Notch pathway was upregulated in ISC and TA subtypes (Fig. 3e, f; Supplementary information, Fig. S5g, h).
To better understand the changes of signaling in ISCs and TA cells, we isolated the GFPhigh cells (ISCs) and GFPlow cells (TA cells)33 from the small intestinal crypts of Lgr5-GFP mice (control) and Lgr5-GFP; β7−/− mice and performed bulk RNA-seq. There were no significant changes in the percentage of GFPhigh or GFPlow population between control and β7−/− mice (Fig. 4a). Compared with cells from control mice, the Wnt target genes Myc, Sox9 and the secretory lineage transcription factor Math1 showed decreased expression in both GFPhigh and GFPlow cells from β7−/− mice, while the Notch target gene Hes1 showed significantly increased expression in GFPlow cells (Fig. 4b). Gene set enrichment analysis (GSEA) showed consistent results, including enrichment of Wnt signature genes in GFPhigh and GFPlow cells from control mice, and enrichment of Notch target genes in cells from β7−/− mice (Fig. 4c). Moreover, the cell cycle genes were significantly enriched in GFPlow cells in β7−/− mice (Fig. 4c), which is consistent with the increased proliferation of crypt cells observed in β7−/− mice (Fig. 2b). Furthermore, we observed consistent changes of Wnt and Notch signal pathways in the crypt cells in β7−/− mice including an increased level of active Notch intercellular domain (NICD), decreased amount of total β-catenin and a slight decrease in active β-catenin (Fig. 4d, e). The immunohistochemical staining showed fewer nuclear β-catenin+ IECs (Fig. 4f) and more HES1+ and NICD+ IECs in the crypts of β7−/− mice as compared with WT mice (Fig. 4g; Supplementary information, Fig. S6). Through analysis of the frequency of nuclear β-catenin+ and HES1+ cells appearing at specific positions in the crypts, we found upregulated Notch signaling and downregulated Wnt signaling along the crypt axis in β7−/− mice (Fig. 4h).
a FACS plots (left) and frequency quantification (right) of GFPhigh and GFPlow cells isolated from control mice (Lgr5-GFP) and β7−/− mice (Lgr5-GFP; β7−/−). n = 3 mice per group, data are represented as means ± SEM, ns, not significant, t-test. b Normalized RNA-seq expression values of Wnt and Notch pathway target genes in GFPhigh and GFPlow cells from control mice (Lgr5-GFP) and β7−/− mice (Lgr5-GFP; β7−/−). n = 5 mice per group, data are represented as means ± SEM, ****P < 0.0001, ***P < 0.001, *P < 0.05 (Deseq2 P values). c GSEA of Wnt, Notch pathway and cell cycle-associated genes in GFPhigh (top) and GFPlow (bottom) cells (control vs β7−/−). The green line shows the enrichment profile. The black bars show where genes from a given gene set are located (hit). NES, normalized enrichment score; FDR, false discovery rate. FDR < 0.25 was considered as statistically significant. d, e Immunoblot analysis (d) and quantification (e) for NICD, β-catenin and active β-catenin in the lysates of the small intestinal crypts from WT and β7−/− mice. n = 3 mice per group. Data are represented as means ± SEM, *P < 0.05, t-test. f Left, representative images of β-catenin staining for crypt sections from WT and β7−/− mice. Nuclear β-catenin+ cells are indicated by red arrows. Scale bars, 25 μm. Right, quantification of nuclear β-catenin+ cells in each crypt. n = 6 mice per group, more than 8 fields per mouse. Data are represented as means ± SEM, *P < 0.05, t-test. g Left, representative images of HES1 staining of crypt sections from WT and β7−/− mice. Scale bars, 25 μm. Right, quantification of HES1+ cells in each crypt. n = 6 mice per group, more than 8 fields per mouse. Data are represented as means ± SEM, *P < 0.05, t-test. h Frequency of nuclear β-catenin+ (middle) and HES1+ (bottom) cells at each position in the small intestinal crypt according to the scheme in the inset (top). n = 4 mice per group, more than 30 well-oriented crypts in each mouse were analyzed. Data are represented as means ± SEM, **P < 0.01, *P < 0.05, ns, no significant difference, t-test.
The data above suggest that the crypt-residing lymphocytes are important for maintaining Wnt and Notch signaling activities in ISCs and TA cells in the small intestinal crypts.
Adoptive T cell transfer rescues aberrant IEC differentiation in β7 −/− mice
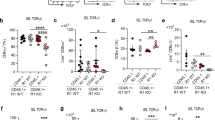
To dissect the role of crypt-residing T and B lymphocytes in regulating IEC differentiation, we transferred WT CD3+ T cells or CD19+ B cells into β7−/− mice to restore the T or B cell population in the small intestine (Supplementary information, Fig. S7a). One week after lymphocyte transfer, we found that transfer of T but not B cells rescued the skewed differentiation of Paneth cells, goblet cells and enterocytes (Fig. 5a), reinforcing that gut-residing T cells regulate IEC differentiation. Furthermore, we profiled the transcriptome of 10,581 IECs using scRNA-seq from the β7−/− mice that received T cell transfer (n = 3 mice). Unsupervised clustering showed that the proportions of the differentiated cell types in the recipient β7−/− mice were rescued after T cell transfer (Fig. 5b). Transfer of β7−/− T cells did not rescue the skewed IEC differentiation in β7−/− mice (Supplementary information, Fig. S7b). These results were further confirmed using Rag1−/− mice, which lack T and B lymphocytes. The Rag1−/− mice showed similar IEC differentiation defects to those in β7−/− mice (Supplementary information, Fig. S8). Adoptive transfer of T cells but not B cells rescued the skewed differentiation of Paneth cells, goblet cells and enterocytes in Rag1−/− mice (Supplementary information, Fig. S8). Moreover, depletion of T cells in WT mice by anti-Thy1.2 antibody treatment resulted in IEC differentiation defects similar to those observed in β7−/− mice (Supplementary information, Fig. S9). Taken together, these results indicate an important role of T cells in regulating IEC differentiation.
a T cells and B cells were isolated from WT mouse spleens and then adoptively transferred into β7−/− mice intravenously. Equal volumes of PBS were injected into WT and β7−/− mice as controls. Mice were sacrificed 7 days post-transfer and the small intestine was assessed using immunohistochemical analysis. Left, representative images of LYZ, PAS and AP staining of small intestine sections for each group. Scale bars, 50 μm. Right, quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus. n = 6 mice per group, more than 8 fields per mouse. ***P < 0.001, **P < 0.01, *P < 0.05, ns, no significant difference, one-way ANOVA. b Cluster frequency analysis depicting the fraction of IEC subtypes obtained by scRNA-seq in WT mice and β7−/− mice 7 days after receiving T cell transfer. ScRNA-seq data from three independent replicates are shown. Cluster identities are indicated. Data are represented as means ± SEM, likelihood ratio test. c Representative images of CD4+ (red) and CD8+ (yellow) T cells in the crypts of WT and β7−/− mice (top), and quantification of CD4+ (single positive), CD8+ (single positive), CD4+ CD8+, or CD4-CD8- cells per crypt section (bottom). n = 4 mice per group, more than 8 fields per mouse. Indigo, CD3; blue, DAPI. Scale bars, 50 μm. *P < 0.05, ns, no significant difference, t-test. d WT CD4+ or CD8+ T cells were adoptively transferred into β7−/− mice intravenously, and equal volumes of PBS were injected into WT and β7−/− mice as controls. The small intestine was assessed using immunohistochemical analysis 7 days post-transfer. Left, representative images of LYZ, PAS and AP staining of small intestine sections for each group. Scale bars, 50 μm. Right, quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus. n = 6 mice per group, more than 8 fields per mouse. ***P < 0.001, **P < 0.01, ns, no significant difference, one-way ANOVA. e Quantification of TCRαβ+ and TCRγδ+ T cells in the crypt region of WT mice. f Quantification of naïve (CD62L+ CD44−), effector memory (CD62L-CD44+) and central memory (CD62L+ CD44+) T cells in the crypt region of WT and β7−/− mice. g Quantification of activated marker CD69+ and CD69− T cells in the crypt region of WT and β7−/− mice. h Quantification of CD4+ T helper cells in the crypt region of WT and β7−/− mice. Th1, CD4+ T-bet+; Th2, CD4+ GATA3+; Th17, CD4+ RORγt+; Treg, CD4+ Foxp3+. n = 4 mice per group, data are represented as means ± SEM, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns, no significant difference, t-test.
We next stained T cell subpopulations and found that CD4+ and CD8+ single-positive T cells were major T cell populations that were significantly decreased in the crypts of β7−/− mice (Fig. 5c). Both CD4+ and CD8+ T cells showed direct contact with ISCs or TA cells in the crypts of WT mice (Supplementary information, Fig. S10a, b). Then, we transferred either CD4+ or CD8+ T cells into β7−/− mice and rescued the CD4+ or CD8+ T cell subpopulation in the crypts (Supplementary information, Fig. S10c). Notably, transfer of either CD4+ or CD8+ T cells restored normal IEC differentiation in the small intestine of β7−/− mice (Fig. 5d), suggesting that both CD4+ and CD8+ T cells in the crypts regulate IEC differentiation.
Most of the crypt-residing and crypt-contacted T cells were TCRαβ+ T cells (Fig. 5e). Although there were a few TCRγδ+ cells, these cells did not decrease in β7−/− mice as did the TCRαβ+ T cells (Fig. 5e). Further analysis showed that these crypt-residing and crypt-contacted T cells were mainly CD62L− CD44+ effector memory T cells (Fig. 5f), and most of the crypt-contacted T cells were activated CD69+ T cells (Fig. 5g). Of note, Treg cells were the major T helper cell population in contact with crypt cells (Fig. 5h). In β7−/− mice, these crypt-contacted T cells significantly decreased (Fig. 5e–h).
T cells regulate ISC differentiation through the integrin αEβ7−E-cadherin adhesion axis
To investigate the contribution of T cell–crypt cell contact and T cell-secreted soluble factors in regulating ISC differentiation, we established an intestinal organoid and T cell co-culture system. Intestinal T cells (92.6% integrin αE+) and organoids were mixed and seeded in Matrigel, where T cells can contact the organoids directly (Supplementary information, Fig. S11). To evaluate the contribution of T cell-secreted soluble factors, T cells were added to the outside of the Matrigel to avoid T cell-organoid contact. Mixing T cells and organoids in Matrigel significantly increased LYZ+ Paneth cells and Muc2+ goblet cells (Fig. 6a) and elevated the expression of their marker genes Defa5 and Muc2 in the organoids (Fig. 6b), whereas the enterocyte marker gene Alpi was decreased (Fig. 6b). The proliferation marker genes Ki67 and Ccnb1 were decreased in organoids co-cultured with T cells in Matrigel (Fig. 6b). No differentiation changes were observed when T cells were added to the outside of the Matrigel (Fig. 6a, b). Moreover, RNA-seq of small intestine tissues from WT and β7−/− mice showed no significant changes in the expression of cytokines and chemokines (Supplementary information, Fig. S12), suggesting the defective IEC differentiation in β7−/− mice is not due to the altered cytokine and chemokine expression. Thus, these data strongly suggest that T cells regulate ISC differentiation through T cell–crypt cell contact, rather than T cell-secreted soluble factors.
a, b Co-culture of intestinal organoids with T cells. Freshly isolated intestinal T cells from WT or αE−/− mice were mixed with organoids in Matrigel or added to the medium outside of the Matrigel at a ratio of 1 organoid/100 T cells and co-cultured for 4 days. Organoids cultured without T cells were used as a control. For integrin αE-blocking antibody treatment, isolated intestinal T cells were pre-treated with the integrin αEβ7 mAb M290 (10 μg/mL) and then mixed with organoids at a ratio of 1 organoid/100 T cells in Matrigel followed by 4 days co-culture. a Differentiation markers LYZ for Paneth cells and Muc2 for goblet cells were detected by immunofluorescence. Left, representative images of LYZ+ (red, top panel) cells and Muc2+ (red, bottom panel) cells within co-cultured organoids. Scale bars, 25 μm. Right, quantification of LYZ+ Paneth cells and Muc2+ goblet cells within the organoids for each group. More than 10 images were analyzed per group and data are represented as means ± SEM, ***P < 0.001, ns, no significant difference, one-way ANOVA. b qPCR analysis of lineage and proliferation markers in organoids, including Defa5 for Paneth cells, Muc2 for goblet cells, Alpi for enterocytes, Ki67 and Ccnb1 for proliferation. Data are represented as means ± SEM for triplicate samples, **P < 0.01, *P < 0.05, ns, no significant difference, one-way ANOVA. c Staining of integrin αE+ T cells in the crypts of WT, β7−/− and αE−/− mice. Red, integrin αE; indigo, CD3; green, E-cadherin; blue, DAPI. Integrin αE+ T cells in contact with crypt epithelial cells are indicated by the white arrows. Scale bars, 50 μm. d Left, representative images of LYZ, PAS and AP staining of small intestine sections from WT mice treated with αEβ7 mAb (M290) or control IgG. Scale bars, 50 μm. Right, quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus. n = 4 mice per group, more than 8 fields were analyzed per mouse. **P < 0.01, t-test. e Top, representative images of LYZ, PAS and AP staining of small intestine sections from WT and αE−/− mice. Scale bars, 50 μm. Bottom, quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus. n = 6 mice per group, more than 8 fields per mouse. ***P < 0.001, t-test. f CD4+ and CD8+ T cells were isolated from the spleens of integrin αE−/− mice and adoptively transferred into β7−/− mice intravenously. Equal volumes of PBS were transferred into WT and β7−/− mice as controls. The small intestine was assessed using immunohistochemical analysis 7 days after T cell transfer. Top, representative images of LYZ, PAS and AP staining of small intestine sections for each indicated group. Scale bars, 50 μm. Bottom, quantification of LYZ+ Paneth cells, PAS+ goblet cells and AP+ enterocytes in each crypt or villus. n = 6 mice per group, more than 8 fields per mouse. ***P < 0.001, **P < 0.01, one-way ANOVA.
Lymphocyte–IEC contact was mainly mediated by adhesion between lymphocyte integrin αEβ7 and E-cadherin expressed on IECs.34,35 FACS analysis showed that 61.5% of CD4+ T cells and 98.2% of CD8+ T cells in IELs express αE integrin, and 25.2% of CD4+ T cells and 73.7% of CD8+ T cells in LPLs are αE positive (Supplementary information, Fig. S13a). Notably, T cells in contact with crypt cells were integrin αE positive (Fig. 6c). In WT mice, we observed equivalent αEβ7+ CD4+ and CD8+ T cells in the crypts of small intestine, and similar numbers of αEβ7+ CD4+ and CD8+ T cells in contact with crypt cells (Supplementary information, Fig. S13b). To investigate whether T cells may regulate ISC differentiation via αEβ7−E-cadherin adhesion, we blocked the αEβ7−E-cadherin interaction in the T cell–intestinal organoid co-culture system by using integrin αE knockout (αE−/−) T cells or normal T cells treated with αEβ7-blocking antibody M290.36 Both αE−/− T cells and M290-treated T cells failed to induce the cell differentiation changes as did the normal T cells (Fig. 6a, b), suggesting that the regulation of ISC differentiation by T cells is dependent on αEβ7−E-cadherin adhesion between T cells and crypt cells. These results were further confirmed by blocking αEβ7−E-cadherin interaction in WT mice with M290 mAb or using αE−/− mice, which showed similar defective IEC differentiations as β7−/− mice did (Fig. 6d, e). Furthermore, adoptive transfer of αE−/− CD4+ or CD8+ T cells could not rescue the defective IEC differentiation in β7−/− mice (Fig. 6f), although T cell population was restored to a normal level in the small intestinal crypts after T cell transfer (Supplementary information, Fig. S13c). Similarly, transfer of αE−/− T cells did not rescue the skewed IEC differentiation in Rag1−/− mice (Supplementary information, Fig. S8). These data further support the notion that T cells regulate ISC differentiation through the integrin αEβ7−E-cadherin interaction.
Integrin αEβ7 regulates ISC differentiation via adhesion signaling through E-cadherin
During E-cadherin internalization, E-cadherin-bound β-catenin is released and accumulates in the cytoplasm, and consequently promotes the activation of Wnt signaling.37 Notably, a decreased E-cadherin level on the surface of crypt epithelial cells where contact with T cells was observed in small intestine, suggesting internalization of E-cadherin in crypt cells upon interacting with αEβ7 on T cells (Fig. 7a; Supplementary information, Fig. S14). To investigate the role of αEβ7 in E-cadherin internalization and the consequent signaling, we treated human colon cancer cells HCT116 with conditioned medium containing soluble recombinant human integrin αEβ7 protein (rh-αEβ7). Immunoblotting results showed that rh-αEβ7-containing medium significantly induced the internalization of the membrane-bound E-cadherin and β-catenin (Fig. 7b) and a decrease in NICD and Hes1 levels (Fig. 7c), which could be inhibited by blocking endocytosis with clathrin inhibitor. These results indicate that αEβ7 promotes Wnt signaling and suppresses Notch pathway by inducing the endocytosis of E-cadherin. In addition, the TOPFlash reporter assay showed that enhancement of Wnt signaling by integrin αEβ7 is dependent on Wnt3a (Fig. 7d), which is highly expressed in the crypt region.
a Representative image (left) shows the reduction of E-cadherin on the plasma membrane of crypt epithelial cells contacted with T cells. Green, Lgr5; red, CD3; white, E-cadherin; blue, DAPI. Red arrows indicate the contact interface between epithelial cell and T cell. Scale bars, 25 μm. The graph (right) shows the quantification of E-cadherin intensity on crypt cell membrane regions contacted or uncontacted with T cells. n = 80 crypt sections, data are represented as means ± SEM, ****P < 0.0001, paired t-test. b Western blot analysis (top) and quantification (bottom) of E-cadherin and β-catenin in membrane and cytoplasmic fractions. Membrane and cytoplasm fractions of human HCT-116 cells were collected following incubation with conditioned medium of soluble rh-αEβ7 or control conditioned medium for the indicated time. For clathrin inhibitor treatment, cells were preincubated with 50 μM Pitstop2 for 1 h followed by incubation with rh-αEβ7 conditioned medium. The integrin β1 and GAPDH were used as loading controls for membrane and cytoplasm, respectively. Data are represented as means ± SEM, **P < 0.01, *P < 0.05, ns, no significant difference, one-way ANOVA. c Western blot analysis (top) and quantification (bottom) of active Notch intercellular domain (NICD) and Hes1. Whole cell lysate of HCT-116 cells was collected following treatment as in panel (b). GAPDH was used as loading controls. Data are represented as means ± SEM, ***P < 0.001, **P < 0.01, ns, no significant difference, one-way ANOVA. d Top-flash activity in mouse epithelial cells treated with purified rm-αEβ7 protein. Eph4 cells were transiently transfected with the Top-flash plasmid. After 24 h, cells were incubated with rm-αEβ7 protein and Wnt3a at indicated concentration for 6 h, and then luciferase activity was measured. Data are represented as means ± SEM for 5 replicates per group, ****P < 0.0001, ns, no significant difference, t-test. e, f Intestinal organoids were cultured with or without rm-αEβ7 protein (10 μg/mL) and M290 antibody (10 μg/mL) for 4 days, followed by qPCR analysis for target genes of Wnt (Axin2, Sox9), Notch (Hes1) and proliferation markers (Ki67, Ccnb1) (e) and differentiated IEC markers (f). Data are represented as means ± SEM for triplicate samples, **P < 0.01, *P < 0.05, ns, no significant difference, one-way ANOVA. g Representative images of LYZ and Muc2 staining for organoids cultured with or without rm-αEβ7 protein and M290 antibody for 4 days. Scale bars, 25 μm. h qPCR analysis of differentiated IEC marker genes for organoids treated with rm-αEβ7 protein (10 μg/mL), Wnt inhibitor XAV-939 (10 μM) and Notch activator VPA (1 mM) as indicated for 4 days. Data are represented as means ± SEM, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns, no significant difference, t-test.
To further confirm the role of integrin αEβ7 in the regulation of Wnt and Notch signaling and ISC differentiation, we added purified recombinant mouse integrin αEβ7 (rm-αEβ7) protein to the intestinal organoids. Consistently, the rm-αEβ7 protein induced the upregulation of Wnt target genes Axin2 and Sox9, and the downregulation of Notch target gene Hes1 as well as cell proliferation genes Ki67 and Ccnb1 (Fig. 7e). Consequent effects on IEC differentiation in intestinal organoids were also observed, including upregulation of Paneth and goblet maker genes (Defa5 and Muc2), downregulation of enterocyte maker gene Alpi and the increased expression of lysozyme and Muc2 proteins upon rm-αEβ7 treatment (Fig. 7f, g). Blocking the integrin αEβ7 and E-cadherin interaction with antibody M290 abolished these changes in the gene and protein expressions (Fig. 7e–g). Furthermore, either inhibition of Wnt signaling by XAV-939 or activation of Notch signaling by valproic acid (VPA) abolished the rm-αEβ7-induced upregulation of Defa5 and Muc2 expression, and downregulation of Alpi expression (Fig. 7h). These data indicate that integrin αEβ7 regulates ISC differentiation via adhesion signaling through E-cadherin.
Aberrant antimicrobial peptide expression and bacterial dysbiosis in the gut of β7 −/− mice
Gut microbiota is important for human health.38,39,40,41 Paneth cells are the primary producers of antimicrobial peptides (AMPs) in the intestinal tract, which have important roles in gut microbiota homeostasis and host defense.42 In mice, AMPs mainly include cryptdins (also named α-defensins), lysozyme and angiogenin 4 (ANG4). We next analyzed the expression of cryptdin4, Lyz1 and Ang4, and observed a significant reduction of these AMPs in the gut of β7−/− mice (Fig. 8a). In addition, quantification of typical pathogenic bacteria showed an increased abundance of Escherichia coli, Staphylococcus aureus and Salmonella typhimurium in the gut of β7−/− mice (Fig. 8b). Adoptive T cell transfer restored the levels of AMPs and pathogenic bacteria back to normal in β7−/− mice (Fig. 8a, b). These results indicate that the defective IEC differentiation in β7−/− mice leads to insufficient production of AMPs and the consequent dysbiosis of intestinal microbiota.
a qPCR analysis of relative AMP expression in the small intestine of mice. b Quantification of the indicated bacteria species in the small intestine of mice by qPCR with primers specific to 16 S rDNA genes. n = 4 mice per group, data are represented as means ± SEM, ***P < 0.001, **P < 0.01, *P < 0.05, one-way ANOVA.
Discussion
Immune cells have been recently recognized as important components of ISC niche. Most of these studies mainly focus on the functions of immune cell-derived cytokines under pathological conditions, such as tissue damage and inflammatory response in the gut.22,23,24,27 However, the role of immune cells in regulating gut homeostasis under normal physiological condition is much less understood. Notably, it has long been observed that lymphocytes often directly interact with IECs under normal condition, however, whether and how these lymphocyte–ISC interactions may affect gut homeostasis is largely unknown. Here, we describe a previously unrecognized role of spatial relationships and interactions between gut-residing integrin αEβ7+ T cells and stem/TA cells in regulating ISC differentiation through T cell–stem/TA cell contact-induced adhesion signaling.
Our results support a cell–cell contact model of crosstalk between T cells and stem/TA cells in which T cells impact the ISCs differentiation through cell contact-dependent αEβ7–E-cadherin adhesion signaling. Using an intestinal organoid system, we have shown that this regulation requires T cell–crypt cell contact, rather than soluble T cell-secreted factors, suggesting that αEβ7+ T cells near the ISC compartment can modulate the biology of ISCs independently of their classical immune functions. Different from T cells exerting their immune effects on ISCs by secreting cytokines or cytotoxic factors to regulate damage repair and tissue regeneration under pathological conditions, this kind of regulation might play a more important role in maintaining gut homeostasis under normal physiological condition. Furthermore, the purified αEβ7 integrin protein showed a similar function as αEβ7-expressing CD4+ and CD8+ T cells in regulating signaling pathways that control ISC differentiation, indicating this mechanism is independent of T cell signature. Whether αEβ7–E-cadherin adhesion is the only mechanism that T cells utilize in this process remains to be determined.
Gut epithelium consists of several distinct differentiated cell types.32 The major cell type is enterocyte and a small number of other types of individual cells locate at different intervals. It is tempting to speculate that the selective differentiation of ISCs into these minority cell types requires precise regulation at single cell level. Our model of T cell–crypt cell contact allows the regulation of ISC differentiation at single cell resolution, which may have an important role in lineage determination of these minority cell types.
Integrin αEβ7 is expressed by a variety of immune cells including intraepithelial T cells, dendritic cells and regulatory T cells.43 By binding to its ligand E-cadherin on epithelial cells, αEβ7 contributes to mucosal specific retention of lymphocytes within epithelia. In the gut, almost all IELs and about half of T lymphocyte population in the lamina propria express αEβ7. However, αEβ7 was expressed by few if any B cells in the gut mucosa or elsewhere,43 which is consistent with our data that B cells showed almost no contact with the IECs and could not rescue the aberrant IEC differentiation due to the lack of αEβ7. Moreover, we observed neither αE+ dendritic cells (CD11c+ CD3−) nor the contact between dendritic cells and the epithelial cells in the crypt region, and the αE+ cells contacted with the crypt cells were mainly T cells (Supplementary information, Fig. S15). Furthermore, the contacted T cells in the crypt were mainly TCRαβ+ T cells (Fig. 5e). According to the review by Olivares-Villagomez et al.,44 TCR+ IELs can be further divided into induced and natural IELs. Induced IELs are mainly TCRαβ+ T cells. Natural IELs include TCRαβ+CD8αα+ T cells, TCRγδ+ T cells and TCR- IELs. Although TCRγδ+ T cells are also αEβ7+, these cells are not the majority of T cells in the crypt region (Fig. 5e). Considering our data showing that the contacted T cells in the crypt were mainly TCRαβ+ T cells and the TCRαβ+CD8αα+ T cells are the minority in TCRαβ+ IELs,45 it is tempting to speculate that the induced IELs play a major role in the regulation of ISC differentiation via interacting with E-cadherin on crypt cells.
Ligand binding to integrin may induce the activation of integrin downstream signaling, thus regulating lymphocyte functions.46,47 To investigate whether E-cadherin binding to integrin αEβ7 may induce T cell activation, we treated the mouse intestinal αEβ7+ T cells with mouse E-cadherin protein. The results showed that the binding of E-cadherin to T cells could not induce the increased expression of T cell activation markers CD25, PD-1 and CD69 as did CD3/CD28 mAb stimulation (Supplementary information, Fig. S16), indicating that E-cadherin binding to integrin αEβ7 does not induce T cell activation.
Wnt signaling is a key regulator of cell proliferation and differentiation and involves proteins that directly participate in both gene transcription and cell adhesion.48,49 The central player in Wnt pathway is β-catenin, which is a transcription cofactor with T cell factor/lymphoid enhancer factor (TCF/LEF).50 In addition, β-catenin can associate with the cytoplasmic domain of E-cadherin and act as an essential component of the adherent junction at the membrane.51,52,53 Disrupting E-cadherin/β-catenin complex affects not only the epithelial cell junction but also the Wnt/β-catenin signaling.54 E-cadherin may suppress β-catenin/TCF-mediated transcriptional activity by sequestrating β-catenin into E-cadherin cell-adhesion complexes. However, this process is independent of E-cadherin adhesion function.55,56 In our study, we showed a new function of E-cadherin in regulating Wnt/β-catenin and Notch pathways, which is mediated by the adhesion between integrin αEβ7 and E-cadherin. Of note, our result showed that the activation of Wnt signaling by integrin αEβ7 through binding to E-cadherin required Wnt3a (Fig. 7d). It is reported that Wnt ligands are enriched in the intestinal crypts but not in the villus.15,57 Therefore, T cell αEβ7 and epithelial cell E-cadherin interaction may only regulate the Wnt signaling in ISCs and TA cells in the crypt region which has high concentrations of Wnt but has no such effect on IECs in the villus region which has insufficient Wnt ligands.
Wnt signaling activation is required for secretory progenitor lineage determination.11,21 Notably, the differentiation of enteroendocrine lineage is independent of Wnt activation although enteroendocrine cells also belong to the secretory lineage.58 Consistent with these studies, we found that the decreased Wnt activation in β7−/− crypt epithelial cells resulted in increased number of enteroendocrine cells and decreased number of Paneth and goblet cells. Similarly, another study has also shown that β7−/− mice have an increased number of enteroendocrine cells.59
Taken together, our study demonstrates that integrin αEβ7-expressing T cells regulate the fate decisions of ISCs through an integrin αEβ7−E-cadherin adhesion axis. Integrin αEβ7 on T cells binds to E-cadherin on ISCs and TA cells and then activates Wnt signaling and suppresses Notch signaling in those cells in the crypts, thus controlling IEC homeostasis. These findings highlight the role of T cell–crypt cell contact in ISC fate decisions and the involvement of adhesion signaling. Notably, the mechanism of T cell–crypt cell contact allows the precise spatial regulation of ISC differentiation at the single-cell level. In addition to maintaining IEC homeostasis under normal biological conditions, this T cell–crypt cell contact may also play profound roles in the pathogenesis of intestine diseases.
Materials and methods
Experimental animals
Wide-type (WT) mice, Itgb7−/− (β7−/−) mice, Itgae−/− (αE−/−) and Rag1−/− mice were obtained from Jackson Laboratory. Lgr5-EGFP-IRES-creERT2 (Lgr5-GFP) mice and Rosa26-confetti mice were obtained from Y.A.Z’s Lab. All mice were maintained under specific pathogen-free conditions. Age- and sex-matched mice were used for all experimental procedures at 8–10 weeks of age. Littermates with the indicated genotypes were used for the animal experiments. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Center for Excellence in Molecular Cell Science (CEMCS), CAS.
Immunohistochemistry staining and analysis
Mouse intestine samples were fixed with 4% paraformaldehyde (PFA) (Sigma) and paraffin embedded using a standard histological protocol. 4-μm-thick serial sections were used for the H&E staining and immunohistochemistry staining. Antigen retrieval was performed with Antigen Unmasking Solution (Vector Laboratories) and the detection was performed with a DAB detection kit (Vector Laboratories). Antibodies used were polyclonal rabbit anti-human lysozyme (1:2000, Dako A0099), rabbit anti-β-catenin (1:1600, Cell Signaling Technology 9587), rabbit anti-Hes1 (1:6400, Cell Signaling Technology 11988). PAS and alkaline phosphatase (AP) staining were performed with Periodic Acid-Schiff kit (Sigma) and BCIP/NBT substrate (Sigma) according to the manufacturer’s instructions. Slides were counterstained with hematoxylin (Thermo Fisher Scientific). Measurements for each quantitative outcome were collected from 50–80 crypts or villi each mouse analyzed from more than 8 independent fields of ileum. The position of nuclear β-catenin+ and HES1+ cells were quantified from more than 30 well-oriented crypts for each mouse.
Immunofluorescent staining and microscopic imaging
Dissected mouse intestines were fixed in 4% PFA in PBS for 1 h. The fixed intestines were incubated in cryoprotective solution (30% sucrose) at 4 °C overnight and cryopreserved in OCT (Thermo Fisher Scientific) and stored at −80 °C. 8-μm-thick cryosections were cut using a freezing microtome (Leica). The frozen sections were permeabilized and blocked in PBS containing 0.3% Triton X-100, 5% BSA (Yeasen Biotech) and 5% normal goat serum (MultiSciences) for 1 h at room temperature. Then sections were incubated with the indicated primary antibody overnight at 4 °C, followed by incubation with corresponding secondary antibodies for 2 h at room temperature. Antibodies used were rat monoclonal Alexa Fluor 594 anti-CD3 (1:300, BioLegend 100240), rat monoclonal APC anti-CD19 (1:200, BioLegend 115522), rat monoclonal FITC anti-CD103 (integrin αE) (1:300, BD Biosciences 557494), rat monoclonal eFluor 570 anti-CD4 (1:300, eBioscience 41-0042), rat monoclonal eFluor 660 anti-CD8 (1:300, eBioscience 50-0081), rabbit monoclonal anti-E-cadherin (1:300, Cell Signaling Technology 3195), rabbit monoclonal anti-Fabp1(1:300, Cell Signaling Technology 13368), polyclonal rabbit anti-Mucin2 (1:300, Santa Cruz 15334), polyclonal rabbit anti-ChgA (1:300, Abcam 15160), polyclonal rabbit anti-human lysozyme (1:500, Dako A0099), Armenian hamster monoclonal FITC anti-TCRβ (1:300, eBioscience 11-5961-82), Armenian hamster monoclonal PE anti-TCRγδ (1:300, Biolegend 118108), rat monoclonal PE anti-CD62L (1:100, BD Biosciences 553151), rat monoclonal Cy7 anti-CD44 (1:100, BD Biosciences 561859), Armenian hamster monoclonal APC anti-CD69 (1:100, BD Biosciences 560689), rat monoclonal APC anti-Foxp3 (1:50, eBioscience 17-5773-82), rabbit monoclonal anti-T-bet (1:200, Cell Signaling Technology 14307), rat monoclonal eFluor 660 anti-GATA3 (1:100, eBioscience 50-9966-42), rat monoclonal APC anti-RORγt (1:100, eBioscience 17-6988-82), Armenian hamster monoclonal APC anti-CD11c (1:200, eBioscience 17-0114-82), rabbit monoclonal anti-cleaved Notch1 (NICD) (1:50, Cell Signaling Technology 4147), Alexa Fluor 488-, Alexa 555-, Alexa 647-conjugated secondary antibodies (1:500, Invitrogen). Nuclei were counterstained with DAPI (Invitrogen). Images were captured with an SP8 confocal microscope (Leica) or an FV-1200 fluorescence microscope (Olympus).
For 3D-reconstruction of the crypt regions, small pieces of intestine were tissue cleared using polyethylene glycol (PEG)-associated solvent system (PEGASOS). The whole-mount immunohistochemical staining was performed as previously reported60 and confocal microscope z-stacks were captured using Sp8 confocal microscope (Leica).
For visualization of T cells in the cocultured organoids, T cells were labeled with fluorescence using CellTracker Orange dyes (ThermoFisher C34551) before coculture experiments. Images were captured with an IX73 microscope (Olympus).
For the organoid immunostaining, whole organoids were collected by gently dissolving the Matrigel in ice-cold Cell Recovery Solution (Corning 354253) and subsequently fixed 30 min in 4% PFA. Next, organoids were permeabilized and blocked in PBS containing 0.5% Triton X-100 and 5% normal goat serum for 1 h at room temperature. Organoids were incubated overnight at 4 °C in blocking buffer containing primary antibodies. Primary antibodies used were polyclonal rabbit anti-Mucin2 (1:300, Santa Cruz 15334), polyclonal rabbit anti-human lysozyme (1:500, Dako A0099). Organoids were incubated with Alexa 488- or 555- conjugated anti-rabbit secondary antibodies (1:1000, Invitrogen) in blocking buffer containing DAPI (1:1000, Invitrogen) for 2 h at room temperature. Images were captured with an SP8 confocal microscope (Leica).
Lymphocyte quantification for the crypt section
To quantify the number of T or B cells in the crypt region, we analyzed the immune fluorescence images of intestine sections by staining E-cadherin and indicated lymphocyte markers including CD3, CD19, CD4 and CD8. In each field which contained about 4–12 crypts, we counted the number of the crypts as well as the total number of T or B cells that reside in the lamina propria and the epithelium in the crypt region. The number of T or B cells per crypt section was calculated as the total number of T or B cells divided by the number of crypts in each field. More than 5 fields per mouse were analyzed and the data were collected from 4–5 mice for each group.
To quantify the T cell phenotypes in the crypt region, we analyzed the immune fluorescence images of intestine sections with the indicated lymphocyte markers including TCRαβ, TCRγδ, CD69L, CD44, CD69, T-bet, GATA3, RORγt, Foxp3. Indicated antibodies were used for immunofluorescence staining. The number of T cells that reside in the lamina propria and the epithelium in the crypt region was counted.
EdU incorporation
EdU was injected intraperitoneally into WT and β7−/− mice at 80 mg/kg for 2 h before tissue collection. For EdU staining, frozen sections were stained and imaged using Click-iT EdU Imaging Kit (Thermo Fisher Scientific, C10339).
Crypt isolation and cell dissociation
The crypts were isolated and enriched as described previously.32 In brief, the small intestine was dissected, flushed with cold PBS, opened longitudinally and cut into small fragments roughly 2–4 mm in length. Intestine fragments were washed twice with cold PBS and then incubated with 20 mM EDTA-PBS on ice for 30 min. The tissue was then shaken vigorously, and the supernatant was collected as fraction 1 in a new conical tube. The tissue was incubated in fresh EDTA-PBS and a new fraction was collected every 30 min. Fractions were collected until the supernatant consisted almost entirely of crypts. The final fraction was filtered through a 70-μm cell strainer (enriched for crypts), washed twice in PBS, centrifuged at 300 g for 3 min, and dissociated with TrypLE Express (Invitrogen) at 37 °C for 5 min. The single-cell suspension was then passed through a 40-μm cell strainer.
Cell sorting
For scRNA-seq experiments, purified crypts were enriched from the small intestine of WT and β7−/− mice and dissociated into single cells. The cells were stained for 7AAD− (Life Technologies), CD45−, CD31−, TER-119−, EpCAM+ (eBioscience), and sorted by a BD AriaIII cell sorter. Cells were pelleted by centrifugation at 4 °C and resuspended in 50 μL 0.4% BSA-PBS on ice until proceeding to the GemCode single-cell platform. To enrich stem cells and TA cells for bulk RNA-seq, cells isolated from Lgr5-GFP mice and Lgr5-GFP; β7−/− mice were stained with antibodies mentioned above and gated on GFPhigh (stem cells) and GFPlow (TA cells).
scRNA-seq library construction
Isolated single cells were processed through the 10× Chromium Single Cell Platform using the GemCode Gel Bead, Chip and Library Kits (V2 and V3, 10× Genomics, Pleasanton) as per the manufacturer’s instructions. In brief, single cells were sorted into 0.4% BSA-PBS. An input of 12,000 single cells was added to each channel with a recovery rate of roughly 6800 cells. The cells were then partitioned into Gel Bead-in-Emulsions (GEMs). GEMs underwent reverse transcription to barcode RNA before cleanup and cDNA amplification. Libraries were prepared with the Chromium Single Cell 3′ Reagent Version 2 or 3 Kit and were further sequenced on an Illumina Hiseq Xten (2 × 150) platforms at the Omics Core of Bio-Med Big Data Center, CAS-MPG Partner Institute for Computational Biology, Shanghai, China.
Analysis of scRNA-seq data
To generate feature-barcode matrices, Illumina basecall files (.bcl) were converted to fastqs and then aligned to the mm10 mouse genome using cellranger v2.1.1 and v3.0.0 according to the library versions. Subsequent cell clustering analyzes were conducted using standard workflow using the R package Seurat.61 First, doublets were removed using doubletFinder software (DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors) for each sample and cells with too few genes detected or high mitochondrial gene-expression fractions were excluded. Then, all samples were merged using merge function from Seurat and were normalized to the same sequencing depth using log normalized method. Top 2000 highly variable genes (HVGs) were calculated using FindVariableFeatures function for PCA analysis. To remove batch effects, we ran the RunHarmony function from R package Harmony62 on the Seurat pipeline using the first 30 PCs. Then, a shared nearest neighbor modularity optimization-based clustering using the FindClusters function with resolution = 1.8 was used to generate all cell clusters. Marker genes were calculated using FindAllMarkers function, and cell clusters were assigned based on known markers of each IEC cell type. The neighborhood graphs were embedded using RunUMAP function and visualized using DimPlot function in Seurat.
Statistic analysis of shifts in cell proportions in vivo
To control variability among mouse-to-mouse biological replicates, we used a negative binomial generalized linear model as previously described.24 In short, the fraction of each cell type was modeled by the total cell number of each mouse as an offset variable, with the genotype of each mouse (i.e., WT or β7−/−) acted as a covariate. The R glm.nb function from MASS package was used for fitting this model. Likelihood-ratio test was used to detect the significance of the effect produced by the β7 knockout.
Stem and TA subsets partitioning
ISCs and TA cells from all cells integrated with WT and β7−/− mice were extracted and re-analyzed using Seurat pipeline as described above. After removing low-quality cells, we obtained three sub-clusters for ISCs and TA cells using resolutions of 0.2 and 0.3, respectively.
Cell–cell similarity matrix
The cell–cell similarities were computed to visualize the sub-clustering heterogeneity of ISCs and TA cells. The first ten principal component scores for each cell were selected to calculate Pearson correlation using R function rcorr which calculates the distance between cells and visualized as a heatmap using the R function pheatmap.
Cell-cycle characteristics of ISC and TA subsets
To identify the cell cycle state for each subset of ISCs and TAs, we downloaded gene sets associated with G1/S and G2/M phases of the cell cycle from https://www.cell.com/cms/attachment/2051395126/2059328514/mmc2.xlsx,63 and re-selected these signature genes with correlation (r > 0.3) of mean expression for each gene in these gene sets of each sub-cluster. The mean expression of genes that are associated with a particular cell-cycle phase in each sub-cluster was calculated.64
Scoring cells using signature gene sets
The AddModuleScore function in Seurat package was used to calculate the average expression of a specific set of genes at a single cell level, number of bins was set default and number of control features selected from the same bin per analyzed feature was set to 5.
Bulk cell and tissue sample RNA-seq
For the bulk population samples, RNA was isolated using the RNeasy Micro Kit (Qiagen) as instructed in the manufacturer’s instructions. For the tissue samples, total RNA was extracted using RNAiso Plus (Takara) from the ileums. The RNA-seq libraries were then proceeded according to the standard Illumina protocols.
Gene set enrichment analysis (GSEA)
GSEA was performed following standard recommended procedures (http://software.broadinstitute.org/gsea/). The Wnt signaling gene set was generated from the Wnt homepage website https://web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes. The Notch signaling gene sets were from the MSigDB C2 collection (REACTOME_SIGNALING_BY_NOTCH.gmt). The cell cycle gene sets were from the MSigDB (KEGG_CELL_CYCLE.gmt). The cytokine and inflammation pathway gene sets were from the MSigDB (BIOCARTA_CYTOKINE_PATHWAY.gmt) (BIOCARTA_INFLAM_PATHWAY. gmt).
Immunoblot analysis
Cells were incubated with conditioned medium of soluble recombinant human αEβ7 (rh-αEβ7) or control conditioned medium for the indicated time. For clathrin inhibitor treatment, cells were preincubated with 50 μM Pitstop2 (GlpBio) for 1 h followed by incubation with rh-αEβ7 conditioned medium. For the whole cell lysate analysis, cells were lysed with lysis buffer (Cell Signaling Technology) supplemented with protease and phosphatase inhibitor cocktail (Roche) on ice. For the subcellular fraction analysis, fractions were prepared using Membrane and Cytosol Protein Extraction Kit (Beyotime). Equal amounts of total protein from each sample were fractionated by SDS-PAGE and blotted onto polyvinylidene difluoride membrane. Protein blots were hybridized with the indicated primary antibody of interest and then with secondary antibody. Immunoblotting was performed with antibodies against E-cadherin (1:2000, Abcam Ab76055), β-catenin (1:2000, Cell Signaling Technology 9587), active β-catenin (1:2000, Cell Signaling Technology 8814), NICD (1:1000, Cell Signaling Technology 4147), Hes1 (1:2000, Cell Signaling Technology 11988). For the whole cell lysate analysis, GAPDH or β-actin was detected by immune blot using anti-GAPDH or anti-β-actin antibody (1:5000, MultiSciences ab011, ab010) as a loading control. For the subcellular fraction analysis, integrin β1 (1: 2000, Abcam Ab52971) and GAPDH antibodies show the membrane and cytoplasmic localization and were also used as loading controls. Fold changes were quantified using ImageJ software.
Intestinal organoid culture
Following crypt isolation, the crypt suspension was resuspended in growth-factor-reduced Matrigel (Corning). Roughly 300 crypts embedded in 50 μL Matrigel were seeded onto each well of a 24-well plate. Once solidified, the Matrigel was incubated in 600 μL culture medium, which is advanced DMEM/F12 medium (Invitrogen) supplemented with 2 mM GlutaMAX, 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen), 10 mM HEPES, 1 mM N-acetylcysteine (Sigma), B27 (1×, Gibco), N2 (1×, Gibco), 50 ng/mL mouse recombinant EGF (Invitrogen), 100 ng/mL mouse recombinant noggin (PeproTech) and 5% R-spondin 1-conditioned medium.
For recombinant mouse integrin αEβ7 (rm-αEβ7) treatment assay, organoids were cultured in the complete culture medium supplemented with or without 10 μg/mL purified rm-αEβ7. The M290 blocking antibodies were added in the culture medium at a final concentration of 10 μg/mL for 30 min at 37 °C before seeding the organoids. Wnt inhibitor XAV-939 (10 μM) and Notch activator VPA (1 mM) were used according to previous studies.65,66 The organoids were cultured for 4 days and then were collected using a cell recovery solution (Corning) on ice for 30 min to dissolve the Matrigel.
Intestinal lymphocytes isolation
Intestinal mucosal lymphocytes were prepared using a modification of a previously published procedure.67 Briefly, the small intestines were removed from mice euthanized in accordance with institutional guidelines. After removal of Peyer’s patches, small intestines were rinsed twice with Ca2+ /Mg2+-free Hank’s balanced salt solution containing 2% FBS (NATOCOR) and then cut into 5 mm-length pieces. Intestinal pieces were shaken in pre-digestion buffer, which is PBS buffer supplemented with 2% FBS, 5 mM EDTA (Sigma), 10 mM HEPES (Sigma) and 4 mM DTT (Sigma) at 37 °C, three times for 20 min at 110 rpm. Cells were collected after each shake and pooled. IELs were collected from the interface of a 40%–70% Percoll gradient (GE Healthcare) following 20 min centrifugation at 1000× g without a break. After removal of the epithelial layer, LPLs were released from the lamina propria after finely chopping intestinal segments, followed by incubation in digestion buffer which is RPMI 1640 medium supplemented with 10% FBS, 0.5 mg/mL collagenase D (Roche), 0.5 mg/mL DNase I (Roche), 3 mg/mL Dispase II (Roche) and 10 mM HEPES for 60 min at 210 rpm at 37 °C. LPLs were isolated by discontinuous Percoll gradient centrifugation as described above. For quantification of intestinal T and B cell number, the isolated cells were stained with anti-CD3 and anti-CD19 antibodies to assess T and B cell populations by flow cytometry using a BD Calibur flow cytometer. For preparation of T cell co-culture experiments, intestinal T cells were isolated by negative selection using EasySep mouse cell isolation kit (StemCell Technologies).
T cell co-culture experiments
Organoids were co-cultured with freshly isolated intestinal T cells from WT or αE−/− mice at a ratio of 1 organoid/100 T cells. T cells were added either to the medium or suspended in Matrigel. To elevate the contact of T cells with organoids, we pellet T cells and organoids by centrifugation at 4 °C, 850× g for 20 min. To block the binding of αEβ7 and E-cadherin, T cells were incubated with αEβ7 mAb (M290) (10 μg/mL) for 30 min at 37 °C in advance and then were mixed with organoids in the Matrigel after precipitation and resuspension. Matrigel and culture medium were both supplemented with M290 antibodies (10 μg/mL). Cells were co-cultured for 4 days. The organoids were collected by gently dissolving the Matrigel in ice-cold Cell Recovery Solution (Corning 354253), centrifuged at 4 °C, 150× g for 2 min, repeated twice to remove T cells and debris, followed by qPCR assays.
QPCR analysis
Total RNA was extracted using RNAiso Plus (Takara) from the organoids or tissues. RNA was reverse transcribed with Reverse Transcriptase M-MLV or PrimeScriptTM RT Master Mix (Takara). QPCR was performed using an SYBR Premix ExTaq kit (Takara) on QuantStudio™ 6/7 Flex machine (Applied Biosystems). The comparative threshold cycle method and internal control (GAPDH) were used to normalize the expression of the target genes. Primers for the genes are available in Supplementary information, Table S1.
Protein expression and purification
Recombinant human and mouse αEβ7 proteins with full length ectodomains were purified as described.68 Briefly, soluble integrin was purified from the culture supernatant of 293T cells stably expressing soluble integrin αEβ7 ectodomains with C-terminally fused His tag and Strep-tag II using Ni-NTA agarose (Qiagen) followed by Strep-Tactin (IBA) affinity chromatography and gel filtration (Superdex 200; GE Healthcare). Rat monoclonal antibodies (mAbs) against mouse integrin αE (M290) and integrin β7 (Fib504) were generated by standard hybridoma technology.
Adoptive cell transfer
CD3+, CD4+, CD8+ T cells, or B cells were isolated from mouse spleen using EasySep mouse cell isolation kit for each cell type (StemCell Technologies). The isolated cells were verified by flow cytometry with a purity typically > 95%. For WT T cell or B cell transfer, 1 × 107 fresh isolated T cells or B cells from the spleen of WT mice were resuspended in 200 μL PBS and then adoptively transferred into β7−/− or Rag1−/− mice intravenously. For αE−/− CD4+, CD8+ or CD3+ T cell transfer, 3 × 107 isolated T cells from the spleen of αE−/− mice were transferred into β7−/− or Rag1−/− mice. Immunohistology analysis was performed 7 days after cell transfer.
In vivo antibody administration
WT mice were treated with 200 μg anti-αEβ7 (M290), anti-β7 (Fib504), anti-Thy1.2 or IgG2a control (BioXcell) mAbs per mouse via intraperitoneal injection every other day for two weeks and then were harvested on day15.
TOP-flash reporter assay
TOP-flash reporter assays were performed as described previously.69 Mammary epithelial Eph4 cells were seeded in 96-wells and transfected with 25 ng TOP plasmid, 25 ng GFP plasmid and 150 ng LacZ plasmid using Lipo2000 (Life Technologies) and treated with Wnt3a and rm-αEβ7 for 6 h. Cell extracts were collected for luciferase assay using Luciferase RGA high sensitive kit (Roche). The luciferase activities were analyzed and normalized to GFP expression using a BioTek Synergy NEO machine.
Cell adhesion assay
96-well assay plates (Corning) were coated with mouse E-cadherin-Fc (40 μg/mL) in 50 μL/well coating buffer, overnight at 4 °C. 1% BSA was used to determine background cell adhesion in the absence of E-cadherin. The wells were subsequently washed twice with coating buffer and then blocked with 1% BSA for 1 h at 37 °C. Then the plate was washed with Hanks’ balanced salt solution (HBSS) twice and placed at 4 °C for standby use. Freshly isolated intestinal T cells were labeled with 1 mg/mL BCECF-AM (Molecular Probes) for 15 min at 37 °C. The cells were subsequently washed twice with washing buffer (5 mM EDTA in HBSS) and HBSS. Adhesion assays were carried out in HBSS with combinations of 1 mM CaCl2 and MgCl2. In antibody blocking experiments, cells were preincubated with mAbs for 10 min at 4 °C. Then, 1 × 105 cells/well were incubated for 30 min at 37 °C, and nonadherent cells were removed by washing with HBSS. Fluorescence was determined using the Envision multi-labeled microplate reader (Perkin Elmer). The excitation filter centered at 485 nm, and the emission centered at 535 nm.
Quantification of intestinal bacteria by qPCR
Fecal DNA was extracted from the fecal pellets of indicated mice with a TIANamp Stool DNA kit (TIANGEN DP328-02) according to the manufacturer’s instructions. Quantitative PCR analysis was performed using an SYBR Premix ExTaq kit (Takara) on QuantStudio™ 6/7 Flex machine (Applied Biosystems). Absolute bacterial abundance was determined using standard curves produced from serial dilutions of cloned bacterial DNA corresponding to the short segment of the 16S rDNA gene that was amplified using specific primers as described previously.70 The relative abundance of bacteria in the gut was determined by dividing the taxon-specific 16S rDNA gene copy number by the universal 16S rDNA gene copy number. qPCR measures 16S rDNA gene copies per sample. The taxon-specific and universal 16S rDNA gene primers are available in Supplementary information, Table S1.
T cell activation analysis
Intestinal T cells were plated on the immobilized E-cadherin-Fc or anti-CD3/CD28 (BD Biosciences 555273, 553294) and cultured for 48 h. The activation phenotype of CD4+ and CD8+ T cells was determined by the percentage of CD25+ (BD Biosciences 553071), PD-1+ (Biolegend 109104) and high expression of CD69+ (BD Biosciences 560689) cells using flow cytometry.
Statistical analysis
Unless otherwise specified in the figure legend, statistical significance was determined by two-tailed Student’s t-test or one-way ANOVA with Dunnett post-tests using GraphPad Prism software (version 7.0). The resulting P values are indicated as follows: ns, no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data represent the means ± SEM of at least three independent experiments. The P value for the significance of cell proportions in vivo was assessed using a likelihood-ratio test computed using the R function “anova”. The P value for the significance of Wnt and Notch signature score in the scRNA-seq was assessed using Mann-Whitney U-test. For the dot plot graph, each dot point represents one independent biological replicate. For the box whiskers graph, box plot shows the median, box edges represent the first and third quartiles, and the whiskers show minimum and maximum values.
Data availability
ScRNA-seq data have been deposited in National Omics Data Encyclopedia (https://www.biosino.org/node/index) with the accession code OEP000370. Bulk RNA-seq data are deposited in Gene Expression Omnibus with the accession code GSE133691. All other data used in this study are available from the corresponding authors upon reasonable request.
References
Bjerknes, M. & Cheng, H. The stem-cell zone of the small intestinal epithelium. V. Evidence for controls over orientation of boundaries between the stem-cell zone, proliferative zone, and the maturation zone. Am. J. Anat. 160, 105–112 (1981).
Bjerknes, M. & Cheng, H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am. J. Anat. 160, 77–91 (1981).
Barker, N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014).
Meran, L., Baulies, A. & Li, V. S. W. Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cells Int. 2017, 7970385 (2017).
Yan, K. S. et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature 545, 238–242 (2017).
VanDussen, K. L. et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497 (2012).
He, X. C. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 36, 1117–1121 (2004).
Qi, Z. et al. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017).
Clevers, H. & Nusse, R. Wnt/beta-catenin signaling and disease. Cell 149, 1192–1205 (2012).
Farin, H. F., Van, Es, J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529 e7 (2012).
Pinto, D., Gregorieff, A., Begthel, H. & Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17, 1709–1713 (2003).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Degirmenci, B., Valenta, T., Dimitrieva, S., Hausmann, G. & Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018).
Greicius, G. et al. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA 115, E3173–E3181 (2018).
Shoshkes-Carmel, M. et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 (2018).
Fre, S. et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964–968 (2005).
Jensen, J. et al. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44 (2000).
Pellegrinet, L. et al. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240 e1–7 (2011).
Korinek, V. et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383 (1998).
van Es, J. H. et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Sancho, R., Cremona, C. A. & Behrens, A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 16, 571–581 (2015).
Lindemans, C. A. et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015).
Gronke, K. et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320 e22 (2018).
Wagner, N. et al. Critical role for beta7 integrins in formation of the gut-associated lymphoid tissue. Nature 382, 366–370 (1996).
Schon, M. P. et al. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J. Immunol. 162, 6641–6649 (1999).
Fu, Y. Y. et al. T cell recruitment to the intestinal stem cell compartment drives immune-mediated intestinal damage after allogeneic transplantation. Immunity 51, 90–103 e3 (2019).
Cheroutre, H., Lambolez, F. & Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456 (2011).
Chen, Y., Chou, K., Fuchs, E., Havran, W. L. & Boismenu, R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc. Natl. Acad. Sci. USA 99, 14338–14343 (2002).
Dahan, S. et al. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology 140, 550–559 (2011).
Gorfu, G., Rivera-Nieves, J. & Ley, K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr. Mol. Med. 9, 836–850 (2009).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Taraszka, K. S. et al. Molecular basis for leukocyte integrin alpha(E)beta(7) adhesion to epithelial (E)-cadherin. J. Exp. Med. 191, 1555–1567 (2000).
Cepek, K. L. et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature 372, 190–193 (1994).
Roberts, K. & Kilshaw, P. J. The mucosal T cell integrin alpha M290 beta 7 recognizes a ligand on mucosal epithelial cell lines. Eur. J. Immunol. 23, 1630–1635 (1993).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host. Microbe. 14, 195–206 (2013).
Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71 (2021).
Schroeder, B. O. & Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089 (2016).
Wong, S. H. & Yu, J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 16, 690–704 (2019).
Yang, W. & Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol. Immunol. 18, 866–877 (2021).
Bevins, C. L. & Salzman, N. H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011).
Hadley, G. A. & Higgins, J. M. Integrin alphaEbeta7: molecular features and functional significance in the immune system. Adv. Exp. Med. Biol. 819, 97–110 (2014).
Olivares-Villagomez, D. & Van Kaer, L Intestinal intraepithelial lymphocytes: sentinels of the mucosal barrier. Trends Immunol. 39, 264–275 (2018).
Gapin, L., Cheroutre, H. & Kronenberg, M. Cutting edge: TCR alpha beta+ CD8 alpha alpha+ T cells are found in intestinal intraepithelial lymphocytes of mice that lack classical MHC class I molecules. J. Immunol. 163, 4100–4104 (1999).
Dang, N. H., Torimoto, Y., Schlossman, S. F. & Morimoto, C. Human CD4 helper T cell activation: functional involvement of two distinct collagen receptors, 1F7 and VLA integrin family. J. Exp. Med. 172, 649–652 (1990).
Ni, H. T., Deeths, M. J. & Mescher, M. F. LFA-1-mediated costimulation of CD8+ T cell proliferation requires phosphatidylinositol 3-kinase activity. J. Immunol. 166, 6523–6529 (2001).
Cadigan, K. M. & Nusse, R. Wnt signaling: a common theme in animal development. Genes Dev. 11, 3286–3305 (1997).
Brembeck, F. H., Rosario, M. & Birchmeier, W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 16, 51–59 (2006).
MacDonald, B. T., Tamai, K. & He, X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009).
Gumbiner, B. M. & McCrea, P. D. Catenins as mediators of the cytoplasmic functions of cadherins. J. Cell Sci. Suppl. 17, 155–158 (1993).
Adams, C. L., Nelson, W. J. & Smith, S. J. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion. J. Cell Biol. 135, 1899–1911 (1996).
Mareel, M. et al. E-cadherin/catenin/cytoskeleton complex: a regulator of cancer invasion. J. Cell Physiol. 173, 271–274 (1997).
Nelson, W. J. & Nusse, R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303, 1483–1487 (2004).
Gottardi, C. J., Wong, E. & Gumbiner, B. M. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153, 1049–1060 (2001).
Stockinger, A., Eger, A., Wolf, J., Beug, H. & Foisner, R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol. 154, 1185–1196 (2001).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190 e4 (2017).
He, S. et al. Gut intraepithelial T cells calibrate metabolism and accelerate cardiovascular disease. Nature 566, 115–119 (2019).
Jing, D. et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 28, 803–818 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual. Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015).
Kowalczyk, M. S. et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
Davies, M. D. & Parrott, D. M. Preparation and purification of lymphocytes from the epithelium and lamina propria of murine small intestine. Gut 22, 481–488 (1981).
Qi, J. et al. Identification, characterization, and epitope mapping of human monoclonal antibody J19 that specifically recognizes activated integrin alpha4beta7. J. Biol. Chem. 287, 15749–15759 (2012).
Mao, J. et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7, 801–809 (2001).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Acknowledgements
This research was supported by National Key R&D Program of China (2020YFA0509000 to J.C. and 2019YFA0802001 to Y.A.Z.), the National Natural Science Foundation of China (32030024, 31830112, 31525016 to J.C., 31970702 to C.L., and 31625020, 31530045, 31830056, 31861163006, 31661143043 to Y.A.Z.), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12010101 to J.C. and XDB19020200, XDA16020200 to Y.A.Z.), Program of Shanghai Academic Research Leader (19XD1404200), National Ten Thousand Talents Program. The authors gratefully acknowledge the support of SA-SIBS scholarship program. We thank Dr. Lin Li for scientific discussion and providing the plasmids of TOP-flash assay. We also thank the Genome Tagging Project Center, CEMCS, CAS for technical support.
Author information
Authors and Affiliations
Contributions
S.C., Y.Z., and J.C. designed experiments. S.C., Y.Z. and H.D. performed experiments. S.C. and Y.Z. analyzed data. X.R., H.F. and A. Z. analyzed scRNA-seq data. S.C., Y.Z., L.Y., Y.W., C.L., S.W., M.H., Z.Y, D.W., H.W., G.G., A.Z., Y.A.Z, and J.C. interpreted results. J.C. supervised the research. The manuscript was drafted by S.C. and edited by G.G., A.Z., Y.A.Z. and J.C. All authors contribute to the writing and provide advice.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, S., Zheng, Y., Ran, X. et al. Integrin αEβ7+ T cells direct intestinal stem cell fate decisions via adhesion signaling. Cell Res 31, 1291–1307 (2021). https://doi.org/10.1038/s41422-021-00561-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-021-00561-2
This article is cited by
-
MET receptor serves as a promising target in melanoma brain metastases
Acta Neuropathologica (2024)
-
The secreted protein Amuc_1409 from Akkermansia muciniphila improves gut health through intestinal stem cell regulation
Nature Communications (2024)
-
Intestinal cellular heterogeneity and disease development revealed by single-cell technology
Cell Regeneration (2022)